Page path:
- Press Office
- Press releases 2015
- 02.09.2015 Learning from Nature
02.09.2015 Learning from Nature
Learning from Nature: Genomic database standard alleviates search for novel antibiotics
Penicillin and other secondary metabolites
Bremen, 2nd September 2015
Penicillin, an antibiotic discovered by Alexander Fleming in 1928, is well known. While Fleming noticed the effect of this compound by pure chance, nowadays the quest for novel agents relies on systematic research. Meanwhile scientists identified many more secondary metabolites like Erythromycin, an antibacterial drug. The enormous relevance of these natural products in medicine, agriculture and biotechnology is without any doubt. Many living organisms like plants, fungi and bacteria produce these small exotic molecules in several steps of synthesis, and researchers use computer-based methods for novel compounds and consider their potential use. Now an international consortium of scientists published the minimal standards to organize the data in the scientific journal Nature Chemical Biology in order to optimize the quest for novel natural products.
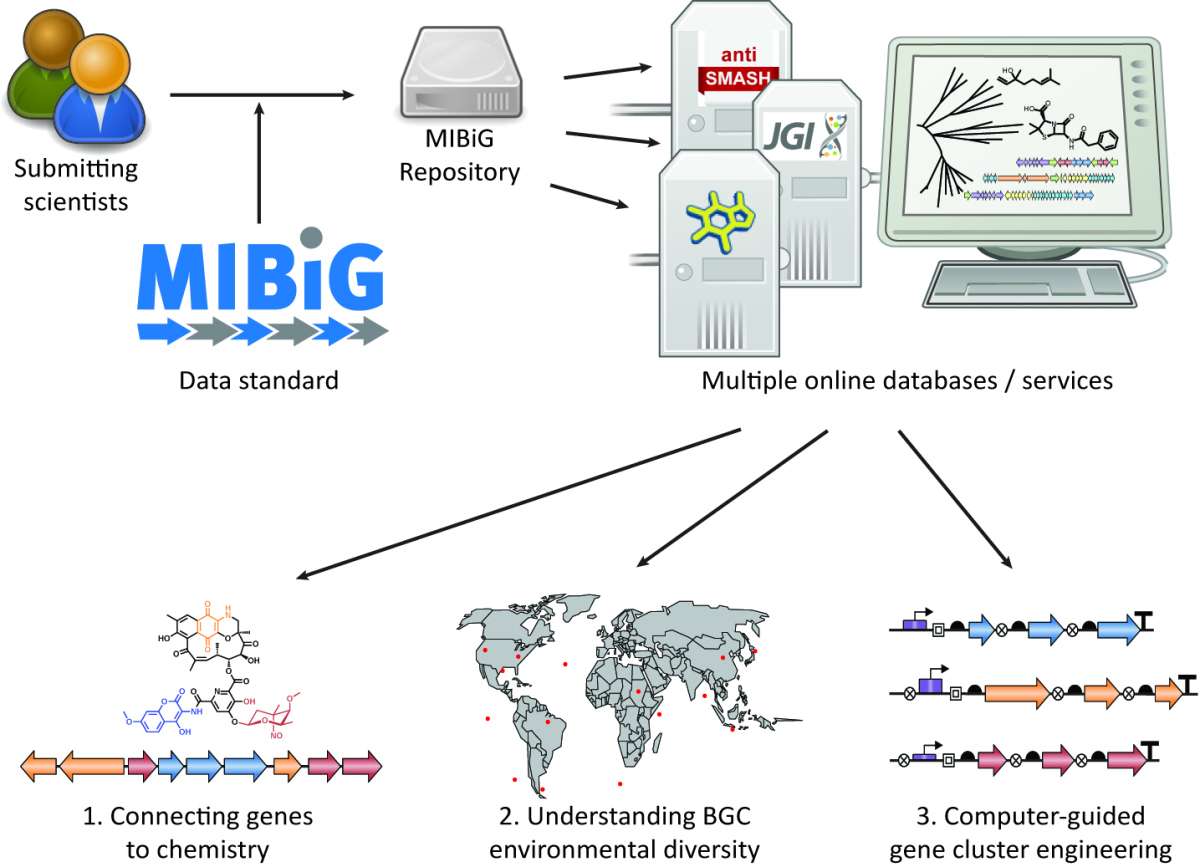
The approach via genome analysis is straightforward and is economical in time and costs as the Biosynthetic Gene Clusters (BGC) can be identified by computer analysis. Only then comes the expensive and time-consuming experimental proof of potential candidate gene clusters in the laboratory. Over ten years ago the Genomic Standard Consortium (GSC) published the first standards for genomic databases.
Prof. Frank Oliver Glöckner – head of the research group Microbial Genomics and Bioinformatics at the Max Planck Institute for Marine Microbiology in Bremen – and his coworker Prof. Marnix Medema have founded this consortium. Glöckner says: “At present the information about these gene clusters is scattered across hundreds of articles in different journals. Without a common standard it is cumbersome to extract, combine and evaluate these data. Our consortium consists of hundreds of scientists, who agreed upon four parameters for each BGC in the database. The main points are who has published what kind of gene cluster in which journal, where is the gene cluster located, what does it produce, and what kind of experimental proof was published. This “Minimal Information about a Biosynthetic Gene Cluster” (MIBiG) links the existing databases and will alleviate the quest for novel substances.” The scientists are sure that with their work it will be much easier to reconstruct the relationships between gene clusters, their chemical potential and their biological diversity.
Penicillin and other secondary metabolites
Bremen, 2nd September 2015
Penicillin, an antibiotic discovered by Alexander Fleming in 1928, is well known. While Fleming noticed the effect of this compound by pure chance, nowadays the quest for novel agents relies on systematic research. Meanwhile scientists identified many more secondary metabolites like Erythromycin, an antibacterial drug. The enormous relevance of these natural products in medicine, agriculture and biotechnology is without any doubt. Many living organisms like plants, fungi and bacteria produce these small exotic molecules in several steps of synthesis, and researchers use computer-based methods for novel compounds and consider their potential use. Now an international consortium of scientists published the minimal standards to organize the data in the scientific journal Nature Chemical Biology in order to optimize the quest for novel natural products.
The approach via genome analysis is straightforward and is economical in time and costs as the Biosynthetic Gene Clusters (BGC) can be identified by computer analysis. Only then comes the expensive and time-consuming experimental proof of potential candidate gene clusters in the laboratory. Over ten years ago the Genomic Standard Consortium (GSC) published the first standards for genomic databases.
Prof. Frank Oliver Glöckner – head of the research group Microbial Genomics and Bioinformatics at the Max Planck Institute for Marine Microbiology in Bremen – and his coworker Prof. Marnix Medema have founded this consortium. Glöckner says: “At present the information about these gene clusters is scattered across hundreds of articles in different journals. Without a common standard it is cumbersome to extract, combine and evaluate these data. Our consortium consists of hundreds of scientists, who agreed upon four parameters for each BGC in the database. The main points are who has published what kind of gene cluster in which journal, where is the gene cluster located, what does it produce, and what kind of experimental proof was published. This “Minimal Information about a Biosynthetic Gene Cluster” (MIBiG) links the existing databases and will alleviate the quest for novel substances.” The scientists are sure that with their work it will be much easier to reconstruct the relationships between gene clusters, their chemical potential and their biological diversity.
For more information please contact
Prof. Dr. Frank Oliver Glöckner
Max Planck Institute for Marine Microbiology, Celsiusstrasse 1, D-28359 Bremen,
Phone: +49 (0)421 2028 – 970, [Bitte aktivieren Sie Javascript]
or the Press officer
Dr. Manfred Schlösser
Max Planck Institute for Marine Microbiology, Celsiusstrasse 1, D-28359 Bremen,
Phone:+49 (0)421 2028 – 704, [Bitte aktivieren Sie Javascript]
Original Publication
Minimum Information about a Biosynthetic Gene cluster. Marnix H Medema, Renzo Kottmann, Pelin Yilmaz et al. Nature Chemical Biology 11, 625–631 (2015) doi:10.1038/nchembio.1890
The complete article is here http://www.nature.com/nchembio/journal/v11/n9/full/nchembio.1890.html
The article cited above is licensed under a Creative Commons Attribution- Non Commercial-ShareAlike 3.0 Unported License. http://creativecommons.org/licenses/by-nc-sa/3.0/.
Prof. Dr. Frank Oliver Glöckner
Max Planck Institute for Marine Microbiology, Celsiusstrasse 1, D-28359 Bremen,
Phone: +49 (0)421 2028 – 970, [Bitte aktivieren Sie Javascript]
or the Press officer
Dr. Manfred Schlösser
Max Planck Institute for Marine Microbiology, Celsiusstrasse 1, D-28359 Bremen,
Phone:+49 (0)421 2028 – 704, [Bitte aktivieren Sie Javascript]
Original Publication
Minimum Information about a Biosynthetic Gene cluster. Marnix H Medema, Renzo Kottmann, Pelin Yilmaz et al. Nature Chemical Biology 11, 625–631 (2015) doi:10.1038/nchembio.1890
The complete article is here http://www.nature.com/nchembio/journal/v11/n9/full/nchembio.1890.html
The article cited above is licensed under a Creative Commons Attribution- Non Commercial-ShareAlike 3.0 Unported License. http://creativecommons.org/licenses/by-nc-sa/3.0/.
Scheme of the workflow. Scientists import their data via online form into the databank. Automatically this information is linked and fed into other data services.
The access is public: http://mibig.secondarymetabolites.org/index.html
The access is public: http://mibig.secondarymetabolites.org/index.html