- Press Office
- Press releases
- Turning a poison into food
Turning a poison into food
Methanogens are microorganisms that produce methane when little or no oxygen is present in their surroundings. Their methane production – for example in the digestive tract of ruminants – is relevant for global carbon cycling, as methane is a very potent greenhouse gas, but can also be used as an energy source to heat our houses.
A toxic base for growth
The object of the study now published in Nature Chemical Biology are two marine heat-loving methanogens: Methanothermococcus thermolithotrophicus (lives in geothermally heated sediments at around 65 °C) and Methanocaldococcus jannaschii (occurs at deep-sea volcanos at around 85 °C). They obtain their cellular energy by producing methane and receive sulfur for growth in form of sulfide, that is present in their environments. While sulfide is a poison for most organisms, it is essential for methanogens and they can tolerate even high concentrations of it. However, their Achilles’ heel is the toxic and reactive sulfur compound sulfite, which destroys the enzyme needed to make methane. Both organisms studied are occasionally exposed to sulfite in their environment, for example, when oxygen enters and reacts with the reduced sulfide. Its partial oxidation results in the formation of sulfite, and thus the methanogens need to protect themselves. But how can they do this?
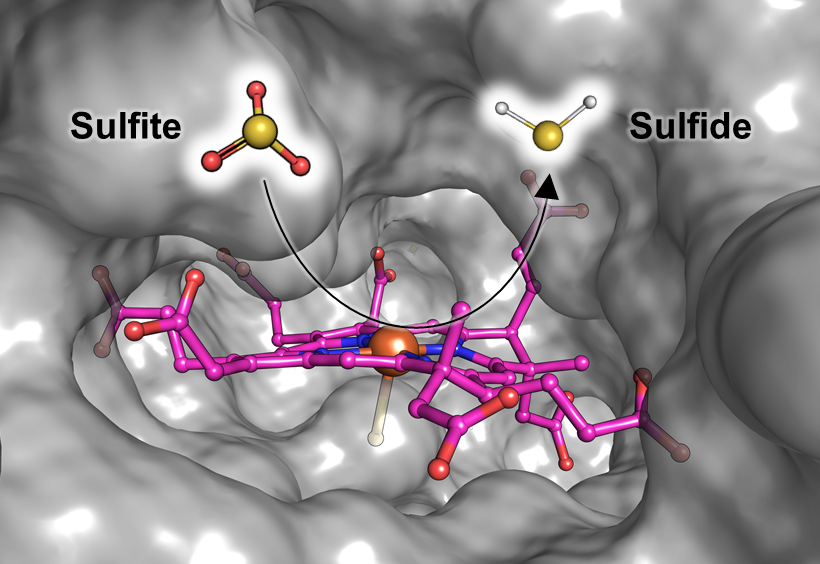
A molecular snapshot of the process
Marion Jespersen and Tristan Wagner from the Max Planck Institute for Marine Microbiology in Bremen, Germany, together with Antonio Pierik from the University of Kaiserslautern, now provide a snapshot of the enzyme detoxifying the sulfite. This butterfly-shaped enzyme ist known as the F420-dependent sulfite reductase or Fsr. It is capable of turning sulfite into sulfide – a safe source of sulfur that the methanogens require for growth. In the current study, Jespersen and her colleagues describe how the enzyme works. “The enzyme traps the sulfite and directly reduces it to sulfide, which can be incorporated, for example, into amino acids”, Jespersen explains (see figure). “As a result, the methanogen doesn’t get poisoned and even uses the product as its sulfur source. They turn poison into food!”
It sounds simple. But in fact, Jespersen and her colleagues found that they were dealing with a fascinating and complicated overlap. “There are two ways of sulfite reduction: dissimilatory and assimilatory”, Jespersen explains. “The organism under study uses an enzyme that is built like a dissimilatory one, but it uses an assimilatory mechanism. It combines the best of both worlds, one could say, at least for its living conditions.”
It is assumed that the enzymes from both the dissimilatory and the assimilatory pathway have evolved from one common ancestor. “Sulfite reductases are ancient enzymes that have a major impact on the global sulfur and carbon cycles”, adds Tristan Wagner, head of the Max Planck Research Group Microbial Metabolism at the Max Planck Institute in Bremen. “Our enzyme, Fsr, is probably a snapshot of this ancient primordial enzyme, an exciting look back in evolution.”

Biotechnological applications in view
The Fsr not only opens up evolutionary implications but also allows us to better understand the fascinating world of marine microbes. Methanogens that can grow only on sulfite circumvent the need to use hydrogen sulfide as a sulfur substrate, which is corrosive, explosive and highly toxic for humans. “This opens opportunities for safer biotechnological applications to study these important microorganisms. An optimal solution would be to find a methanogen that reduces sulfate, which is cheap, abundant, and a completely safe sulfur source”, says Wagner. In fact, this methanogen already exists, it is Methanothermococcus thermolithotrophicus. The researchers hypothesized that Fsr orchestrates the last reaction of this sulfate reduction pathway, because one of its intermediates would be sulfite. “Our next challenge is to understand how the methanogen can transform sulfate to sulfite, to get a complete picture of the capabilities of these miracle microbes.”
Original publication
Jespersen, Marion, Pierik, Antonio J. & Wagner, Tristan (2022): Structures of the sulfite detoxifying F420-dependent enzyme from Methanococcales. Nature Chemical Biology.
Participating institutions
- Max Planck Institute for Marine Microbiology, Bremen, Germany
- University of Kaiserslautern, Kaiserslautern, Germany
Please direct your queries to:
Head of Group
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany

Head of Press & Communications
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
1345 |
|
Phone: |
