Linking “who” and “what”: Protocol for combined (CARD-)FISH and nanoSIMS to visualize specific microorganisms and quantify their activity in the environment
- Departments
- Department of Biogeochemistry
- Biogeochemistry Group
Biogeochemistry Group
Research in the Biogeochemistry Group focuses on microbiological and geochemical processes that control bioactive element cycling in the marine environment. We employ geochemical, microbiological, modeling, molecular and single-cell techniques to study the environmental regulation of these processes, and their effects on the global biogeochemical cycles.
Our goal is to provide fundamental insights into microbial mediated processes in the Ocean that ultimately affect Ocean chemistry, biology and climate, and vital input for models used to predict potential future changes resulting from human activities.
News

20. November 2023
Farooq Jalaluddin successfully defended his PhD thesis - congratulations, Dr. Jalaluddin :)

17. December 2021
Clarissa Karthäuser successfully defended her PhD thesis - congratulations, Dr. Karthäuser :)

3. November 2021
New Paper – Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium
Our latest paper led by Wiebke Mohr is just out in Nature. In the paper we show how a symbiotic marine bacterium provides nitrogen to seagrass. Ca. Celerinatimonas neptuna lives inside the roots of the Mediterranean seagrass Posidonia oceanica and fixes N2 which it provides to the plant during the summer, when nutrient-N concentrations are extremely low in the surrounding seawater. In return for the fixed N, the seagrass likely provides sugars to the bacterium. The metabolic and genomic features of this symbiosis are reminiscent of those that exist between terrestrial N2-fixing microorganisms and their plant hosts. Considering the vast ecological benefits of such a symbiosis, ancestors of the root-endophytic bacterium were likely a key factor which enabled seagrasses to conquer nutrient-poor coastal marine environments, where today they form vibrant ecosystems.
Find our Article, the accompanying News & Views Article by Doug Capone and the Press Release.

13. October 2021
Miriam Philippi successfully defended her PhD thesis - congratulations, Dr. Philippi :)
06. August 2021
New Paper - Purple sulfur bacteria fix N2 in a Proterozoic Ocean analogue
Who knew that the purple waters of Lake Cadagno provide a new perspective on early Earth N2 fixation:
On a field trip in summer 2018, Miriam Philippi and co performed incubation experiments to test N2 fixation by the microbial community in the sulfidic, low molybdenum waters of Lake Cadagno. Suprisingly, purple sulfur bacteria (PSB) were responsible for high N2 fixation rates, and they used an enzyme that was assumed not to be involved in N2 fixation under these low Mo conditions. This finding has important implications for N2 fixation on early earth: PSB were present also in the Proterozoic and based on our results, may have substantially contributed to bioavailable N-supply in photic, sulfidic Proterozoic ocean waters. Previously, this important process for expansion of the early biosphere had been attributed to cyanobacteria.
Read the manuscript at Nature Communications, the corresponding press release and EOS article by Elise Cutts.
Many thanks for a great collaboration with our colleagues from EAWAG and SUPSI!
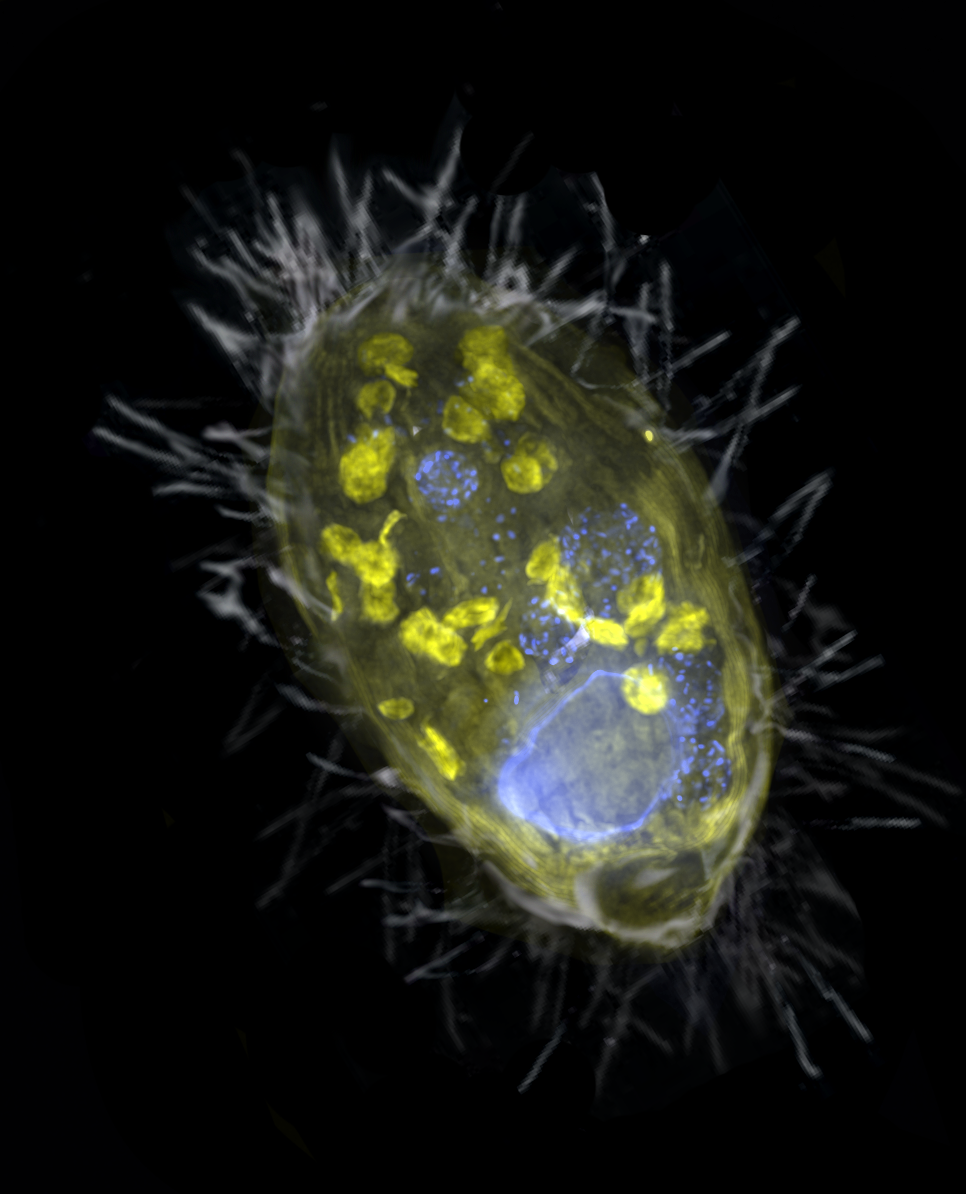
03. March 2021
New Paper - Anaerobic endosymbiont generates energy for ciliate host by denitrification
Our paper on a denitrifying endosymbiont that generates energy for its ciliate host, lead by Jon Graf, and the Greenhouse Gases Research Group, just came out in Nature. We show that an obligate endosymbiont, Candidatus Azoamicus ciliaticola, can respire and provide energy for its host, an anaerobic plagiopylean ciliate. Intriguingly, Azoamicus only encodes the denitrification pathway instead of terminal oxidases, which enables its host to breathe nitrate instead of oxygen. This unprecedented symbiosis raises the possibility that eukaryotes with mitochondrial remnants may secondarily acquire energy-providing endosymbionts to complement or replace functions of their mitochondria.
Read the accompanying News and Views article from Lewis and Ettema and listen to the Nature Podcast where Jana Milucka talks about our discovery.
The press release for our article can be found HERE.
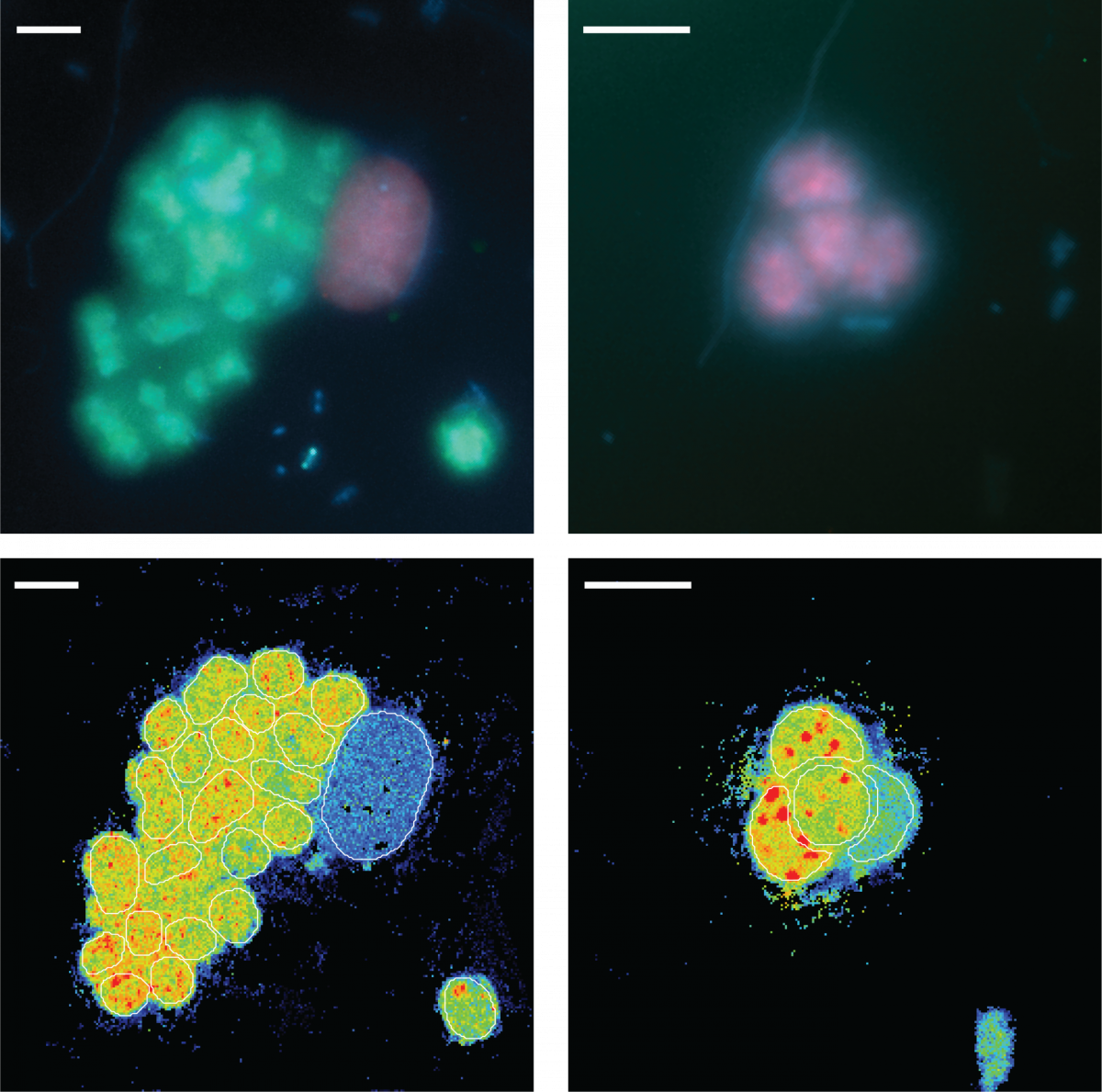
13. February 2021
Catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) is an imaging method used to identify microorganisms in environmental samples based on their phylogeny. CARD-FISH can be combined with nano-scale secondary ion mass spectrometry (nanoSIMS) to directly link the cell identity to their activity, measured as the incorporation of stable isotopes into hybridized cells after stable isotope probing. In environmental microbiology, a combination of these methods has been used to determine the identity and growth of uncultured microorganisms, and to explore the factors controlling their activity. Additionally, FISH-nanoSIMS has been widely used to directly visualize microbial interactions in situ. The biogeochemistry department single cell experts contributed a step-by-step protocol for combined CARD-FISH and nanoSIMS to the book “Fluorescence In-Situ Hybridization (FISH) for Microbial Cells”, part of the Springer Methods in Molecular Biology series (Link).

Congratulations to Julia Dürschlag, who successfully defended her PhD-Thesis (11/20/2020).
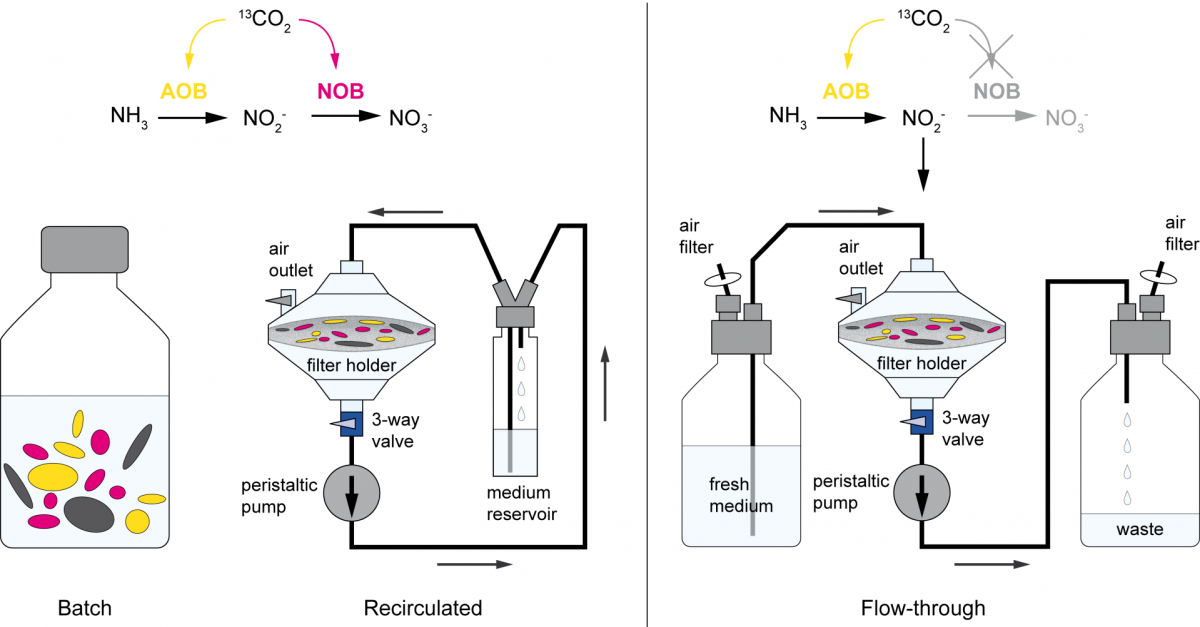
2. September 2020
Are you really what you eat? A new stable isotope probing method to overcome cross feeding
Stable isotope probing (SIP) is a key tool for identifying the microorganisms catalyzing the turnover of specific substrates in the environment and to quantify their relative contributions to biogeochemical processes. However, cross-feeding can pose a serious problem in SIP studies, leading to the erroneous identification of organisms that are not directly responsible for the process of interest, but are rather connected to primary consumers via a microbial food web. In a joint study published in The ISME Journal (Link), colleagues of the University of Vienna, the University of Aalborg, and the MPI for Marine Microbiology describe a new method that rules out cross feeding in SIP studies. In this approach, the microbial cells are exposed on a membrane filter to a continuous flow of medium containing isotopically labeled substrate. Thereby, metabolites and degradation products are constantly removed, preventing consumption of these secondary substrates, thus allowing to distinguish primary consumers from other members of microbial food webs.

Congratulations to one of the pioneers of the Biogeochemistry Group, our technician Swantje Lilienthal...
... on her 25 year anniversary! We are looking forward to the next years.
3. August 2020
The first steps to carbon sequestration in the oceans
In a recent paper in Scientific Reports Hach et al. describe how microbial activity in the oligotrophic surface waters of the North Atlantic Ocean initiate the transformation of dissolved organic matter that is released from phytoplankton.
Dissolved organic matter (DOM) accumulates in the deep ocean because it cannot easily be remineralized to CO2. This so-called recalcitrant DOM represents a major carbon sink and is formed mainly in oligotrophic ocean areas due to microbial breakdown of labile phytoplankton-derived DOM. Despite the fact that DOM export might account for 20% of oceanic CO2uptake, the microbial transformation from labile to recalcitrant DOM is poorly understood. Particularly little is known about the molecular changes associated with this transformation.
In this study (Link), we combined incubation experiments carried out in the North Atlantic ocean with stable isotope tracers and high resolution examination of the DOM pool. Using this novel methodology, we could provide direct experimental evidence showing that within hours of labile DOM release, its breakdown and recombination with ambient DOM leads to the formation of a diverse array of new molecules in oligotrophic North Atlantic surface waters. So far, such molecular diversification was generally believed to be a slow process taking months to years. The resulting diversification of the molecular composition of DOM makes it more resistant to microbial breakdown. The export of this recalcitrant DOM has led to the accumulation of ~662Gt of dissolved organic carbon in the deep ocean. Consequently, the immediate diversification of DOM that already occurs in the surface waters leads to the sequestration of carbon in the deep ocean for millennia, partly counteracting anthropogenic carbon emissions.
24. March 2020
New in Scientific Reports: Golf ball-like sand grains provide ideal structures for microbial colonization
Sandy sediments cover more than 50% of the continental shelf and are akin to biocatalytic-filters, remineralizing carbon and removing nutrients at very high rates. Despite the growing awareness of the importance of sandy sediments in marine nutrient cycling, little is known about the factors that control carbon and nutrient turnover. In a recent publication in Scientific Reports (Link), we show that the surface area of a sand grain that is available for colonization determines microbial cell numbers and is therefore an important factor in controlling the magnitude of respiration rates. Surprisingly, sand grains with a golf-ball like structure are ideal for microbial colonization by providing protective structures, but also by ensuring an optimal solute supply.

Congratulations to Nadine Lehnen and Alexander Khachikyan, who successfully defended their PhD-Theses just before going into lockdown - happy we could still celebrate a little bit! (13/03/2020).
7. February 2020
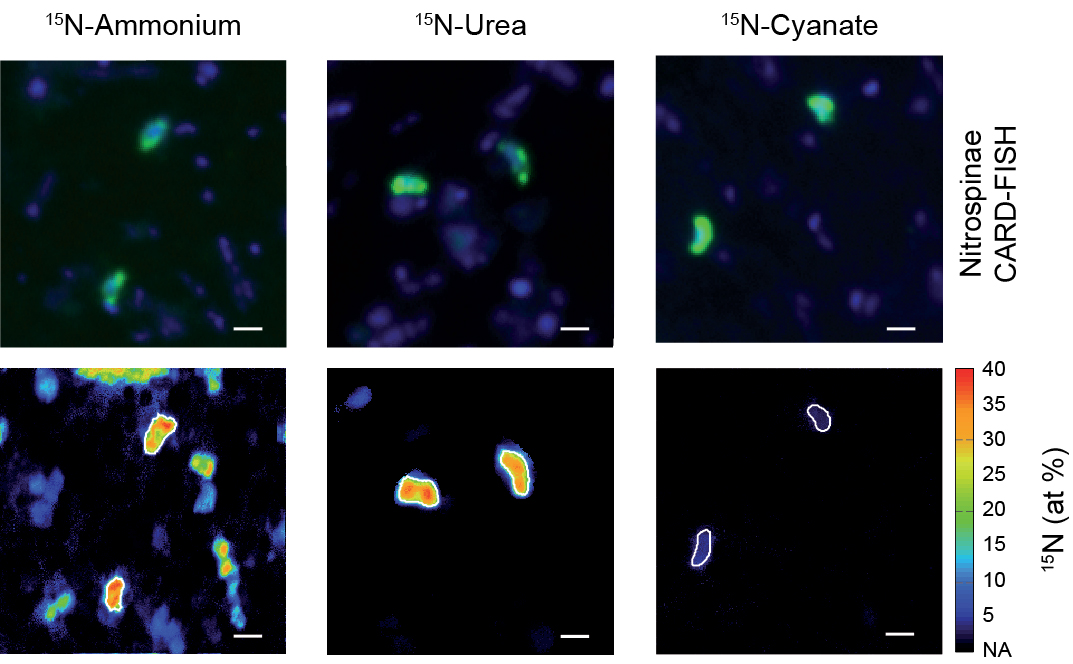
Live fast, die young: Why marine nitrite oxidizers are outnumbered by ammonia oxidizers
Nitrification, the oxidation of ammonia via nitrite to nitrate, is a key process in marine nitrogen cycling. In the sea, both steps of this process are balanced and most available nitrogen exists in the form of nitrate, the final product of nitrification. Despite this, ammonia-oxidizing archaea (AOA) vastly outnumber the main nitrite oxidizers, the bacterial Nitrospinae, though the ecophysiological reasons for this discrepancy in abundance have been unclear. In a new study published in Nature Communications (Link), scientists from the Biogeochemistry Department and colleagues from University of Vienna, the University of Southern Denmark and the Georgia Institute of Technology reveal that differences in mortality between Nitrospinae and ammonia oxidizers, rather than thermodynamics, biomass yield and cell size, determine the abundances of these main marine nitrifiers.