Page path:
- Departments
- Department of Biogeochemistry
- Biogeochemistry Group
- Projects
- Nitrogen and Carbon Assimilation by Marine Microorganisms
Nitrogen and Carbon Assimilation by Marine Microorganisms
In many oceanic regions, primary production is limited by the availability of fixed inorganic nitrogen. The oceanic N-budget is unbalanced with N-loss apparently exceeding N-gain. If real such an unbalance could eventually lead to an end in algal growth. One of our main objectives is to determine if N-gain due to N2-fixation in the Ocean is currently underestimated.
Quantification of nitrogen fixation by individual diazotrophs
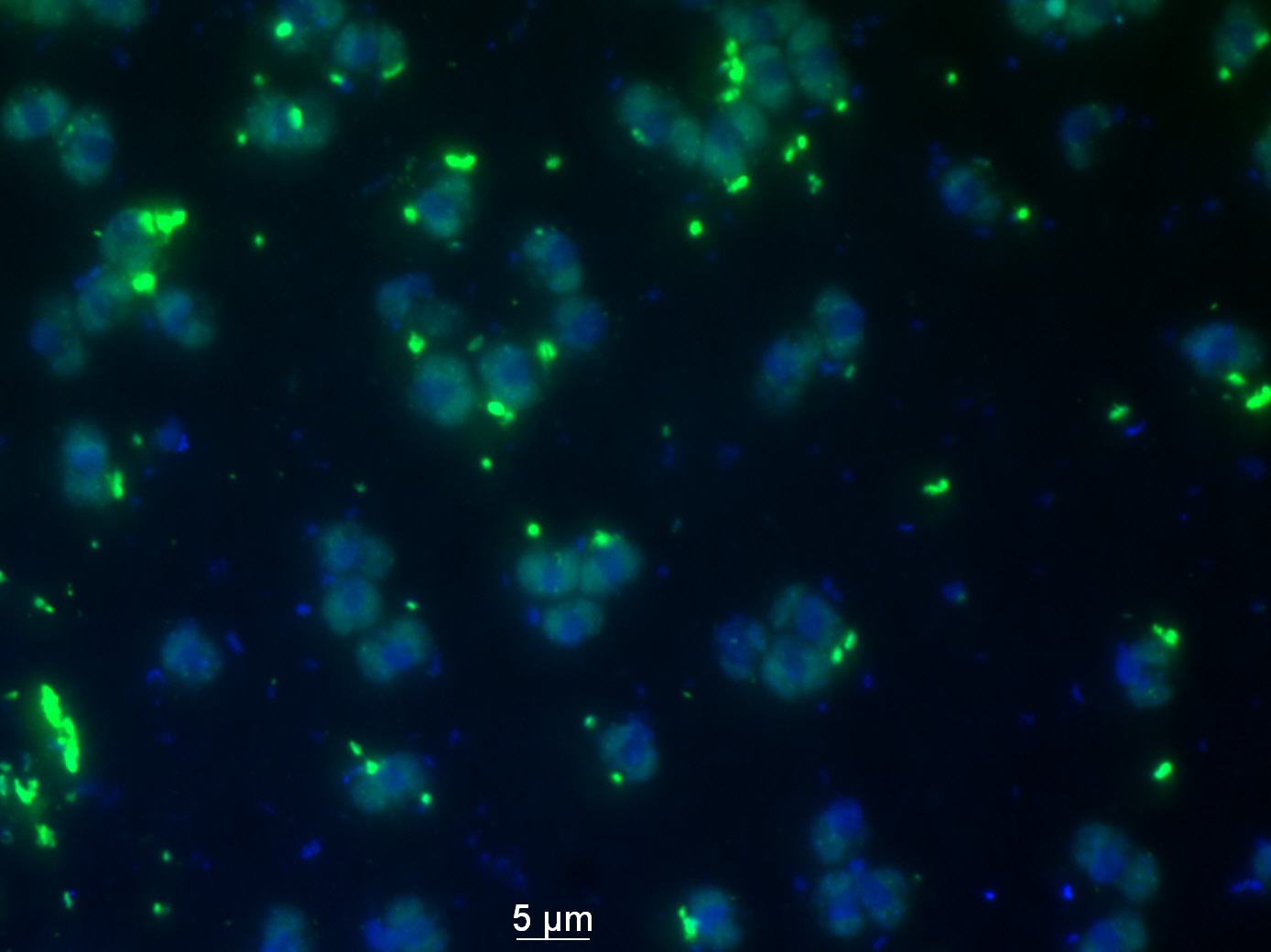
Microorganisms are key players driving the biogeochemical cycling of elements on our planet. As only a very small fraction (< 1%) of extant microorganisms has so far been brought into culture, in situ analysis of microbial diversity and in situ quantification of specific activities of defined microbial populations are crucial in understanding the function of the ‘key players’ in biogeochemical cycling of elements ( Neufeld et al. 2007; Kuypers and Jørgensen, 2007; Amann and Fuchs, 2008; Orphan and House, 2009). In the past years, methods have been developed that combine stable or radioactive isotope tracer experiments to the analysis of DNA, RNA or other biomarkers in order to analyze the function of microorganisms in situ (Dumont and Murrell, 2005; Wagner et al. 2006; Neufeld et al. 2007). These approaches are based on the fact that physiologically active microorganisms in a sample are the ones that incorporate the labeled substrates into cell biomass, DNA or RNA. However, at the moment there are only few approaches with single cell resolution (Neufeld et al. 2007). Recent advances in imaging mass spectrometry have created new research opportunities for the study of single microbial cells in situ (Kuypers and Jorgensen, 2007; Neufeld et al. 2007). NanoSIMS 50, is an instrument with a high lateral resolution (≤ 50 nm in cesium and ≤ 150 nm in oxygen) and enhanced mass-resolving power which has the capacity to measure up to 7 masses (ions) in parallel from the same micro-volume. The high sensitivity and precision of this instrument allows measurements of elements and their isotopes that are present in microbial cells with a precision of the measurement of ±1‰ and thus giving to the nanoSIMS-based methodologies a huge advantage over the existing single cell methods (Kuypers and Jorgensen, 2007; Neufeld et al. 2007; Orphan et al. 2009).
Figure 1 (Musat et al., 2008): 15N-ammonium and 13C-inorganic carbon uptake by individual bacteria inhabiting the chemocline of Lake Cadagno (Switzerland). Parallel secondary ion images of Chlorobium clathratiforme (Cc), Lamprocystis purpurea and Chromatium okenii cells. The abundances of 12C14N (A, E), 19F (B, F), and the 15N/14N (C, G) and 13C/12C (D, H) ratios are shown. Scale bar = 5 m, applicable to all images. Arrows in C indicate highly active epibionts attached to a C okenii cell.
By its ability to measure any stable isotope as well as any radioisotope with a suitable half-life, nanoSIMS can be used to image metabolically active microbial cells within complex communities. Furthermore by coupling of nanoSIMS and in situ hybridization using oligonucleotide probes that are specifically binding to the 16S or 23S rRNA of microorganisms, simultaneous quantification of substrate uptake and phylogenetic identification of microbial cells in a single nanoSIMS scan is possible (Behrens et al. 2008; Li et al. 2008; Musat et al. 2008). The use of HRP-labeled oligonucleotide probes and catalyzed reported deposition of halogenated tyramides makes methods like HISH-SIMS (Halogen In Situ Hybridization-SIMS; Musat et al. 2008; Halm et al., 2009) very useful tools for the in situ analysis of a wide range of environmental samples (Fig.1). The combination of nanoSIMS, in situ experiments with stable isotope labeled substrates and halogen in situ hybridization opens completely new possibilities to pursue other important aspects of the nutrient cycle such as the assimilation of nutrients by prokaryotes. This research will enable us to link the identity of microbial cells in a complex microbial community to their in situ nutrient incorporation (Fig.2), to calculate cellular uptake rates and determine nutrient fluxes resulting from this uptake (Fig.3). Future investigation using this method will for example be used to identify unknown N2-fixers in marine environments and to quantify the contribution of individual N2-fixers to the total nitrogen fixation in these environments.
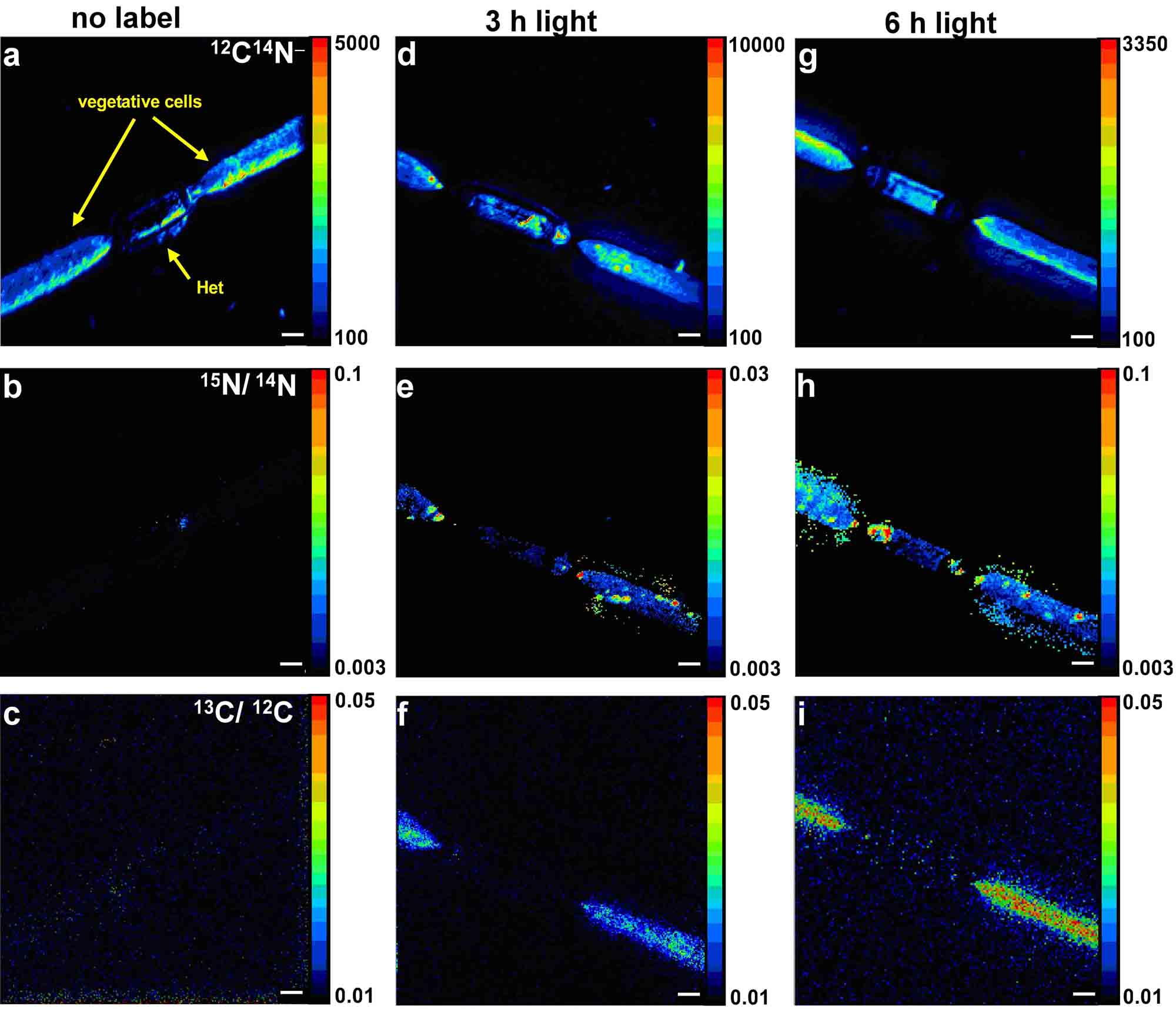
Figure 2 (Ploug, Musat et al., 2010): Distribution of 12C14N- (a,d,g), and 15N/14N (e,h), and 13C/12C (f,i) in single Aphanizomenon cells as measured by nanoSIMS. a-c: no label, d-f: t=3 h, g-i: t=6 h.
The scale bars are 2 microns.
The scale bars are 2 microns.
Effects of viruses on carbon and nitrogen assimilation by microorganisms
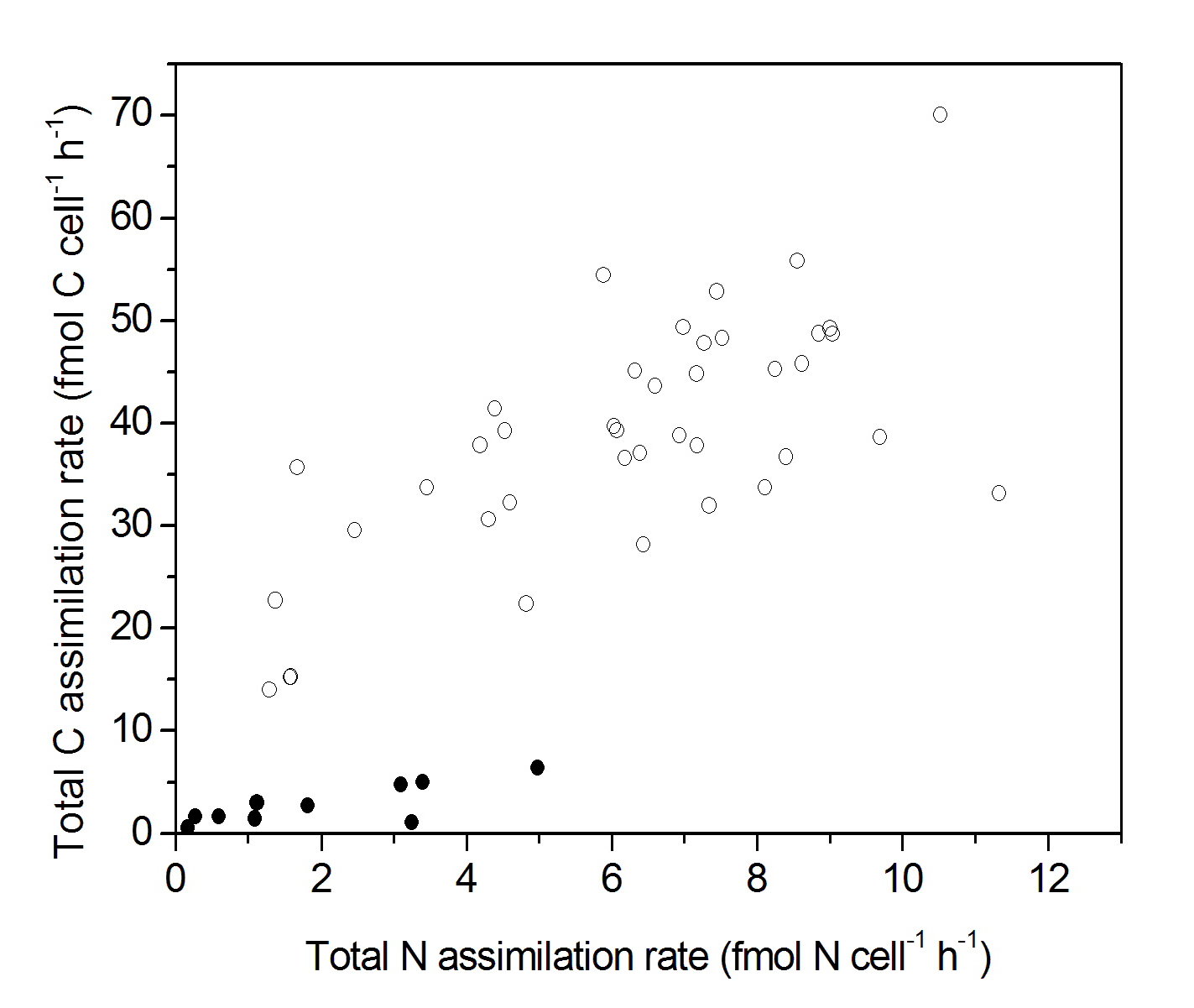
The role of viral lysis on C and N assimilation in marine phytoplankton was investigated in collaboration with the NIOZ, Texel. Incubation experiments with 15N- and 13C-labeled substrates in a phytoplankton culture (Phaeocystis globosa) revealed no significant difference in total C and N assimilation until the point of viral lysis (PgV-07T), though single cell nanoSIMS analyses showed that C and N incorporation by infected algal cells was only ~50% of the incorporation by non-infected cells. These results indicate that viral infection already before viral lysis triggers the exudation of polymeric N-rich organic material by algal cells. Since up to 20% of marine microorganisms maybe infected, these exudates may play an important role in Oceanic C and N cycling.