Page path:
- Departments
- Department of Biogeochemistry
- Biogeochemistry Group
- People
- Research Projects
Research Projects
Molecular Ecology of the Nitrogen Cycle
The global nitrogen cycle is predominantly mediated by microbes, and each of such nitrogen transformations is facilitated by the action of an enzyme. With recent advances in molecular biology, it is now possible to identify the genes encoding these enzymes and to use them as biomarkers to detect the potential or occurrence of these processes in the environment. Table 1 shows some of the representative functional genes that have been successfully applied in environmental studies.
Table 1. Representative functional gene markers for various nitrogen cycling pathways.
| Transformation | Gene(s) | Encoding Enzyme | Reference(s) |
| N2 fixation | nifHDK | Nitrogenase | (Zehr and McReynolds, 1989) |
| Denitrification | narG | Dissimilatory nitrate reductase | (Nogales et al., 2002; Cheneby et al., 2003) |
| nirS, nirK | Nitric oxide reductase | (Braker et al., 2000; Casciotti and Ward, 2001) | |
| norB | Nitric oxide reductase | (Braker and Tiedje, 2003) | |
| nosZ | Nitrous oxide reductase | (Scala and Kerkhof, 1998) | |
| Ammonia oxidation | amo | Ammonia monooxygenase | (Rotthauwe et al., 1997; Alzerreca et al., 1999) |
| hao | Hydroxylamine oxidoreductase | (Hommes et al., 1995; Sayavedra-Soto et al., 1996) | |
| Nitrate assimilation | narB, nasA | Assimilatory nitrate reductase | Allen et al., 2001) |
| Nitrite assimilation | nir | Assimilatory nitrite reductase | Wang et al., 2000) |
| Ammonium assimilation | glnA | Glutamine synthetase | Kramer et al., 1996) |
| Organic N metabolism | ure | Urease | Collier et al., 1999) |
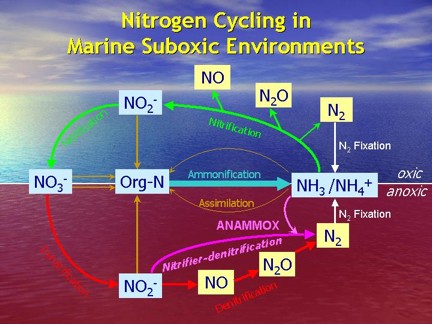
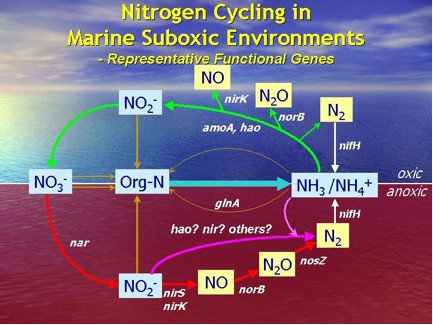
In marine suboxic environment, various microbial nitrogen transformations may be occurring simultaneously that they might be difficult to detect using conventional chemical rate measurements. Therefore, detection of various functional gene markers might be particularly useful in revealing co-occurring processes and complex microbial relationships. On a genetic level, the detection of a particular functional gene shows the potential of that particular process (Fig. 1 & 2). Additional phylogenetic analyses of the functional gene of interest might further elucidate the diversity of the corresponding microbes.
On a level further, the ‘expression’ of the functional gene directly indicates the active building of the encoding enzymes, thus the actual occurrence of the process or function. Currently, our group is particularly interested in the gene expressions of amoA, nirS, nirK and narG, as indicators for aerobic ammonia oxidation, nitrite reduction and nitrate reduction in suboxic environments. Apart from qualitative approaches using reverse transcriptase-polymerase chain reaction (RT-PCR), we would also like to explore how gene expression levels are quantitatively related (via quantitative RT-PCR) to environmental gradients and rate measurements, linking functional genomics to biogeochemical parameters. As the anaerobic ammonium oxidation (or anammox) has been shown an important nitrogen loss pathway in aquatic environments, we are in collaboration with the Department of Microbiology, Radbound University Nijmegen, to identify a suitable functional gene for the anammox process in such functional genomics and gene expression analyses.
On a level further, the ‘expression’ of the functional gene directly indicates the active building of the encoding enzymes, thus the actual occurrence of the process or function. Currently, our group is particularly interested in the gene expressions of amoA, nirS, nirK and narG, as indicators for aerobic ammonia oxidation, nitrite reduction and nitrate reduction in suboxic environments. Apart from qualitative approaches using reverse transcriptase-polymerase chain reaction (RT-PCR), we would also like to explore how gene expression levels are quantitatively related (via quantitative RT-PCR) to environmental gradients and rate measurements, linking functional genomics to biogeochemical parameters. As the anaerobic ammonium oxidation (or anammox) has been shown an important nitrogen loss pathway in aquatic environments, we are in collaboration with the Department of Microbiology, Radbound University Nijmegen, to identify a suitable functional gene for the anammox process in such functional genomics and gene expression analyses.
Figure 1. A simplified nitrogen cycle.
Figure 2. Representative functional genes can be used as biomarkers for various nitrogen transformations in the nitrogen cycle. Please refer to Table 1 for the abbreviations of the functional genes.
References
Allen, A. E., Booth, M. G., Frischer, M. E., Verity, P. G., Zehr, J. P. and Zani, S. (2001). Diversity and Detection of Nitrate Assimilation Genes in Marine Bacteria. Applied and Environmental Microbiology 67(11): 5343-5348.
Alzerreca, J. J., Norton, J. M. and Klotz, M. G. (1999). The amo operon in marine, ammonia-oxidizing [gamma]-proteobacteria. FEMS Microbiology Letters 180(1): 21-29.
Braker, G. and Tiedje, J. M. (2003). Nitric Oxide Reductase (norB) Genes from Pure Cultures and Environmental Samples. Applied and Environmental Microbiology 69(6): 3476-3483.
Braker, G., Zhou, J., Wu, L., Devol, A. H. and Tiedje, J. M. (2000). Nitrite Reductase Genes (nirK and nirS) as Functional Markers To Investigate Diversity of Denitrifying Bacteria in Pacific Northwest Marine Sediment Communities. Applied and Environmental Microbiology 66(5): 2096-2104.
Casciotti, K. L. and Ward, B. B. (2001). Dissimilatory Nitrite Reductase Genes from Autotrophic Ammonia-Oxidizing Bacteria. Applied and Environmental Microbiology 67(5): 2213-2221.
Cheneby, D., Hallet, S., Mondon, M., Martin-Laurent, F., Germon, J. C. and Philippot, L. (2003). Genetic characterization of the nitrate reducing communitz based on narG nucleotide sequence analysis. Microbial Ecology 46: 113-121.
Collier, J., Brahamsha, B. and Palenik, B. (1999). The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme [In Process Citation]. Microbiology 145(2): 447-459.
Hommes, N.-G., Arp, D.-J. and Sayavedra-Soto, L.-A. (1995). Generation of polymerase chain reaction-specific probes for library screening using single degenerate primers. Genetic Analysis Techniques and Applications 12(1): 53-56.
Kramer, J. G., Wyman, M., Zehr, J. P. and Capone, D. G. (1996). Diel variability in transcription of the structural gene for glutamine synthetase (glnA) in natural populations of the marine diazotrophic cyanobacterium Trichodesmium thiebautii. FEMS Microbiology Ecology 21(3): 187-196.
Nogales, B., Timmis, K., Nedwell, D. B. and Osborn, A. M. (2002). Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Applied and Environmental Microbiology 68(10): 5017-5025.
Rotthauwe, J.-H., Witzel, K.-P. and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Applied and Environmental Microbiology 63: 4704-4712.
Sayavedra-Soto, L.-A., Hommes, N.-G., Russell, S.-A. and Arp, D.-J. (1996). Induction of ammonia monooxygenase and hydroxylamine oxidoreductase mRNAs by ammonium in Nitrosomonas europaea. Molecular Microbiology 20(3): 541-548.
Scala, D. J. and Kerkhof, L. J. (1998). Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiology Letters 162(1): 61-68.
Wang, Q., Li, H. and Post, A. F. (2000). Nitrate Assimilation Genes of the Marine Diazotrophic, Filamentous Cyanobacterium Trichodesmium sp. Strain WH9601. Journal of Bacteriology 182(6): 1764-1767.
Zehr, J. P. and McReynolds, L. A. (1989). Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium spp. Applied and Environmental Microbiology 55: 2522-2526.
Allen, A. E., Booth, M. G., Frischer, M. E., Verity, P. G., Zehr, J. P. and Zani, S. (2001). Diversity and Detection of Nitrate Assimilation Genes in Marine Bacteria. Applied and Environmental Microbiology 67(11): 5343-5348.
Alzerreca, J. J., Norton, J. M. and Klotz, M. G. (1999). The amo operon in marine, ammonia-oxidizing [gamma]-proteobacteria. FEMS Microbiology Letters 180(1): 21-29.
Braker, G. and Tiedje, J. M. (2003). Nitric Oxide Reductase (norB) Genes from Pure Cultures and Environmental Samples. Applied and Environmental Microbiology 69(6): 3476-3483.
Braker, G., Zhou, J., Wu, L., Devol, A. H. and Tiedje, J. M. (2000). Nitrite Reductase Genes (nirK and nirS) as Functional Markers To Investigate Diversity of Denitrifying Bacteria in Pacific Northwest Marine Sediment Communities. Applied and Environmental Microbiology 66(5): 2096-2104.
Casciotti, K. L. and Ward, B. B. (2001). Dissimilatory Nitrite Reductase Genes from Autotrophic Ammonia-Oxidizing Bacteria. Applied and Environmental Microbiology 67(5): 2213-2221.
Cheneby, D., Hallet, S., Mondon, M., Martin-Laurent, F., Germon, J. C. and Philippot, L. (2003). Genetic characterization of the nitrate reducing communitz based on narG nucleotide sequence analysis. Microbial Ecology 46: 113-121.
Collier, J., Brahamsha, B. and Palenik, B. (1999). The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme [In Process Citation]. Microbiology 145(2): 447-459.
Hommes, N.-G., Arp, D.-J. and Sayavedra-Soto, L.-A. (1995). Generation of polymerase chain reaction-specific probes for library screening using single degenerate primers. Genetic Analysis Techniques and Applications 12(1): 53-56.
Kramer, J. G., Wyman, M., Zehr, J. P. and Capone, D. G. (1996). Diel variability in transcription of the structural gene for glutamine synthetase (glnA) in natural populations of the marine diazotrophic cyanobacterium Trichodesmium thiebautii. FEMS Microbiology Ecology 21(3): 187-196.
Nogales, B., Timmis, K., Nedwell, D. B. and Osborn, A. M. (2002). Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Applied and Environmental Microbiology 68(10): 5017-5025.
Rotthauwe, J.-H., Witzel, K.-P. and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Applied and Environmental Microbiology 63: 4704-4712.
Sayavedra-Soto, L.-A., Hommes, N.-G., Russell, S.-A. and Arp, D.-J. (1996). Induction of ammonia monooxygenase and hydroxylamine oxidoreductase mRNAs by ammonium in Nitrosomonas europaea. Molecular Microbiology 20(3): 541-548.
Scala, D. J. and Kerkhof, L. J. (1998). Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiology Letters 162(1): 61-68.
Wang, Q., Li, H. and Post, A. F. (2000). Nitrate Assimilation Genes of the Marine Diazotrophic, Filamentous Cyanobacterium Trichodesmium sp. Strain WH9601. Journal of Bacteriology 182(6): 1764-1767.
Zehr, J. P. and McReynolds, L. A. (1989). Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium spp. Applied and Environmental Microbiology 55: 2522-2526.