- Press Office
- Blogs and More
- The census of the seas: Microorganisms and Meiofauna - Blogpost 6 from RV SONNE, March 17, 2018
The census of the seas: Microorganisms and Meiofauna - Blogpost 6 from RV SONNE, March 17, 2018
On Board RV SONNE, Chile
March 17, 2018
Blog #6 The census of the seas: Microorganisms and Meiofauna
How scientists determine marine biodiversity by linking taxonomy and environmental DNA
One teaspoon of sediment contains billions of microbes, each different species well adapted to their small-sized habitat. On a single grain of sand, scientists counted up to 50 000 microbes. While some species prefer the pore water of the sediment, others live in the water column above the seabed. With a size of around one to two micrometers they belong to the unseen majority on our planet. Their biomass accounts for about one third or even more of the total biomass on earth. Despite their size, these tiny cells are in charge of the cycling of the elements on a global scale, as they mineralize the remnants of organic matter and feed carbon dioxide and nutrients back to the cycle.
Moving up one step on the size ladder of organisms, we find the meiofauna. Meiofauna with its strange looking organisms covers the range between 45 micrometers and one millimeter: e.g. ciliates, nematodes and foraminifers. Thanks to their housekeeping action, the blue planet is full of life. Who are the species and how and where are they doing it – that’s one of the fundamental questions for marine scientists.

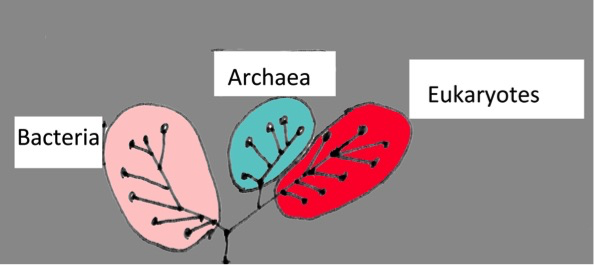
The tree of life
The tree of life reflects the relationship between different species. It can be seen as an ancestor chart with its branches. When finding a new species, taxonomists try to assign its position on this tree and give it a scientific name. To do this, they use several criteria, e.g. the outer shape (morphology) and also certain cell molecules like pigments as markers. This is a well-established method and works fine with larger animals and plants. For tiny sized organisms, it is getting more difficult, as microorganisms come only in a very limited number of shapes, such as spheres, rods and spirals. Also, only a very tiny fraction, much less than 1% of the microbes, can be grown in a lab culture and are thus accessible to detailed analysis. Therefore, scientists have developed cultivation-independent methods based on DNA sequence analysis.
A taxonomic data bank for all species: Hunting for environmental DNA
Many scientists believe there are millions of undetected species out there, and it would take centuries if taxonomy would be done only the classical way. To distinguish one microbial species from another, a light microscope is not sufficient. But there is help from molecular biology. DNA sequences are specific for species. Like in fingerprint analysis on a crime scene, giving the police information about the criminal, marine scientists are applying powerful DNA sequencing techniques when going after their suspects. They are ambitious. Their scientific goal is to categorize every life form, all species living on earth, to complete the tree of life.
Environmental DNA
New tools for investigating environmental DNA (eDNA) are helpful to get an overview. As nature likes to copy success stories, some essential housekeeping genes are conserved with minor differences during evolution and present in every species. In DNA barcoding, a short DNA sequence of a certain gene present in all species under scrutiny is compared. Computer algorithms then calculate the relationship and construct a taxonomic tree.
Metagenomics
With metagenomics it is possible to attempt reconstructing individual genomes. This can be done even in a mixture of several different microorganisms, such as in a typical environmental sample. The first step is to isolate the total DNA from a sediment sample. In the next step, the DNA is artificially amplified in the laboratory by means of the polymerase chain reaction (PCR). Then the scientists sequence the fragments obtained. They feed these DNA sequences into their computers, hoping for overlapping sequences. Then powerful algorithms begin their number crunching.
Basically, this method relies on linguistical methods. It is like comparing a mixture of text fragments from different authors and trying to reconstruct the original texts. Similar to the authors, every species has its different preference of genetic vocabulary as seen in their DNA sequences. Molecular biologists are using their high-end computers to pin down their prey. But as good as this method sounds, it is still a work in progress. To evaluate, improve and to test these DNA based methods, the expert knowledge of taxonomists is vital.
Sophie Arnaud-Haond says: “At Ifremer we are involved in several scientific projects about eDNA. We want to improve the reference databases linking all our new incoming genomic, taxonomic, geographic and environmental data. Therefore, this expedition to the Atacama trench is very important, as here the species can be very different from anywhere else. We want to find out who is adapted to 8000 meters and who are their former relatives living nearby on the abyssal plane.”
Daniela Zeppilli agrees: “We observe similar findings for some meiofauna species. We are interested if nematodes in the deep sea are genetically separated from their ancestors.”
Greetings from the crew and the scientific party of SO261,
Manfred Schlösser
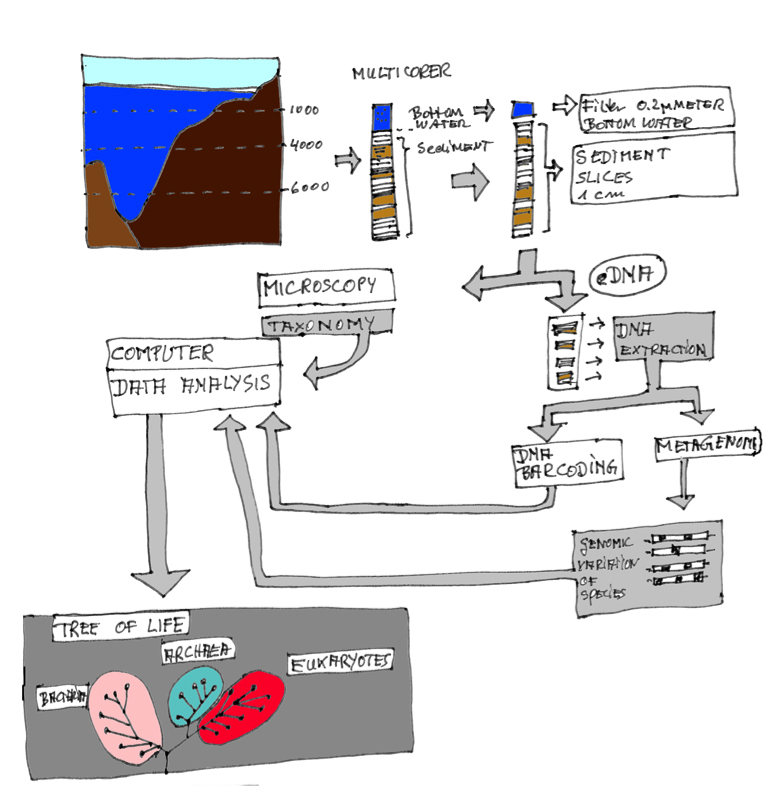
The pipeline for linking taxonomy and DNA data. Taxonomy and genetics are complementary approaches both necessary to describe the biodiversity of the seabed. Sediment samples from the Atacama trench are collected with a multicorer from the seabed in 8000 meters depth. On board RV Sonne, scientists have to work sterile. First they slice the 40 centimeters of the core into one centimeter thick segments. These are fixed and stored in the cold room. A so-called sister core is stored separately and will undergo direct scrutiny by taxonomy. At the end of the pipeline the species are documented in the tree of life.
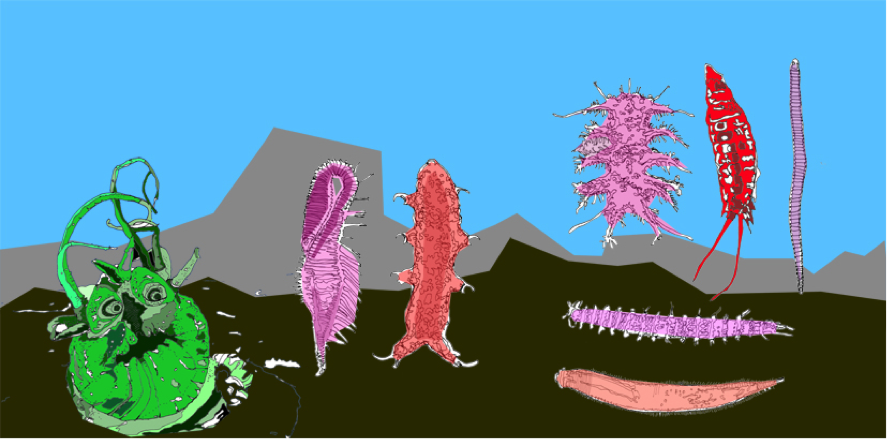
Some meiofauna (size less than one millimeter) found in 8000 meters depth. From left tor right: A head of a polychaete worm, a nematode, two tardigrades (a.k.a. water bears), a kinorhyncha (mud dragon), three nematodes. (Sketches by Manfred Schlösser after photos from Daniela Zeppilli).
Specific questions addressed during this cruise are:
- What are the sedimentary processes providing food for the hadal community in the Atacama Trench?
- How do abundance, diversity and community structure of microorganisms, meio- and macrofauna in the Atacama Trench differ from those in less productive trenches and nearby abyssal and shelf sites?
- What are the general biogeochemical characteristics of the surface and deep sediment and water column in the eutrophic Atacama Trench?
- Which mineralization pathways are responsible for organic matter breakdown in the eutrophic Atacama Trench?
- How efficient are microbial communities operating at extreme hydrostatic pressures in mineralizing organic material as compared to their shallower counterparts? And to what extent do specialized, yet unknown extremophile microbial communities mediate these processes?
Further information
More details about the project from the University of Southern Denmark.
More pictures related to the project.
Ronnie N Glud at Danmarks Radio (in Danish)
RV SONNE is a modern German research vessel sailing mainly in the Pacific Ocean.
More information about the ship here.