- Departments
- Department of Molecular Ecology
- Molecular Ecology
- Fluorescence in Situ hybridization
Fluorescence in Situ hybridization (FISH)
The full rRNA approach and FISH
The full rRNA approach for identification, quantification and localization of yet-uncultured bacteria
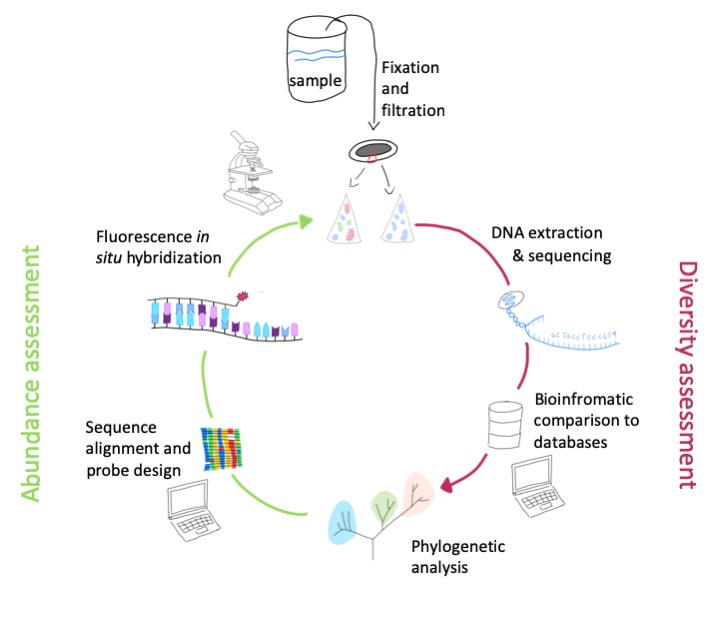
The so-called ribosomal RNA (rRNA) approach has been established more than 30 years ago to study microbial communities. But what is the rRNA approach based on? In contrast to multicellular organisms, unicellular microbes can hardly be distinguished based on their morphology. To make matters worse, not even 1% of all environmental microbes can be cultured in the lab. Instead, the analysis of their genetic material is the right choice, especially studying their rRNA. The rRNA is an essential component of ribosomes, the sites of protein biosynthesis. It is present in every living organism and has approximately the same length in all organisms. It is therefore called "conserved." Comparison of rRNA gene sequences provides information about the relatedness of the organisms from which the rRNA sequences originated. The fewer the differences found in the sequences, the more closely the organisms are related to each other. Thus, it is possible to deduce their ancestry, reconstruct phylogenetic trees and place the microbes in taxonomic order.
With this approach, entire microbial communities can be investigated without having to laboriously culture and characterize the individual members. Additionally, we can study and identify yet-uncultivated and novel organisms.
The schematic workflow can be seen on the right: In the laboratory, the entire DNA is isolated from the environmental sample and the gene segments coding for the rRNA are amplified with specific primers using polymerase chain reaction (PCR). In the next step, the products from the PCR are sequenced and the resulting sequences of the rRNA genes are compared with the already known sequences from public databases. In this way, the diversity of the microbes living in the sample can be comprehensively determined. The next step is the visualization and quantification of the microorganisms, which is described in the next paragraph.
Fluorescence in situ hybridization
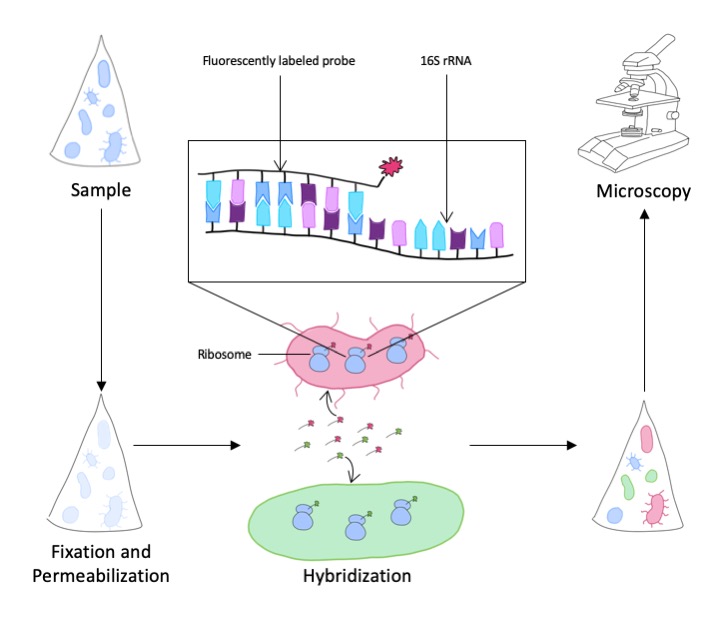
From the above-described rRNA sequence information, we cannot derive the abundance of a particular species or genus in the environmental sample. For this purpose, we use another method to stain and thus visualize and count the microbes in the sample. At first, we have to identify signature sequences in the rRNA that a are specific to the targeted taxonomic group of microorganisms. Next, complementary short DNA sequences labeled with a fluorescent dye, called probes, are synthesized against these signatures. Under suitable experimental conditions, the probes bind to the rRNA of the target organisms only. We can, thus, not only stain but also localize the microorganisms in our samples. This is important, as microbes frequently live in consortia or attached to surfaces.
A prerequisite for the sufficient detection and localization of microorganisms in environmental samples is that they are preserved with a fixative (e.g. formaldehyde) as soon as possible after they have been sampled. The aim is to preserve the natural biocoenosis in the best possible form and distribution.
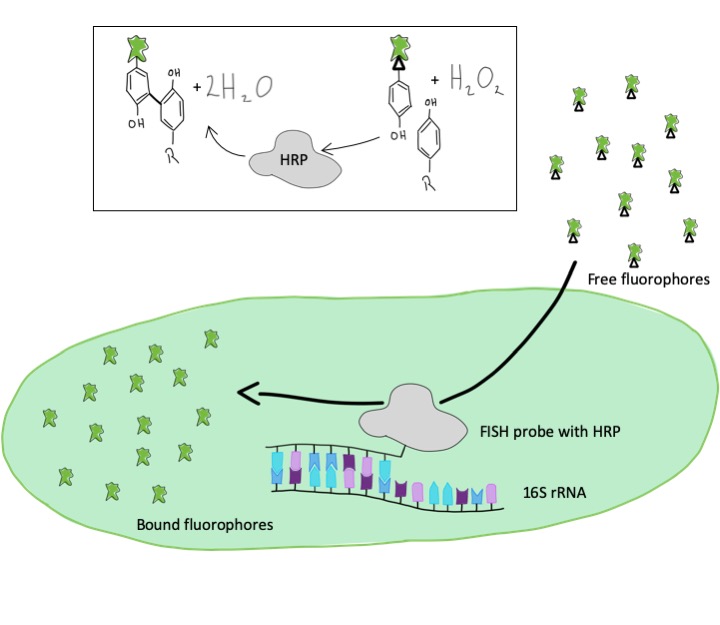
Catalyzed Reporter Decomposition - a stronger signal with CARD-FISH
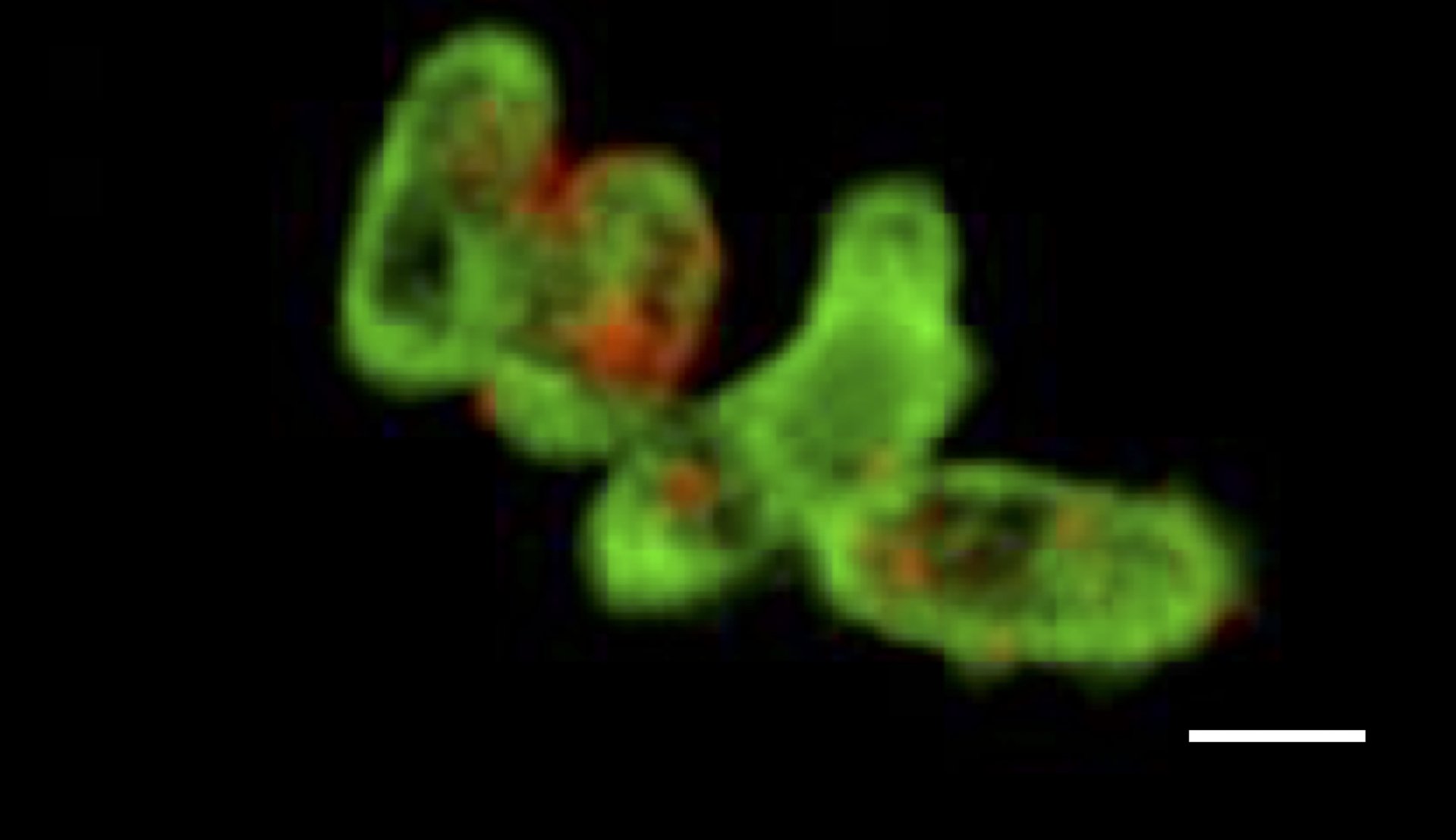
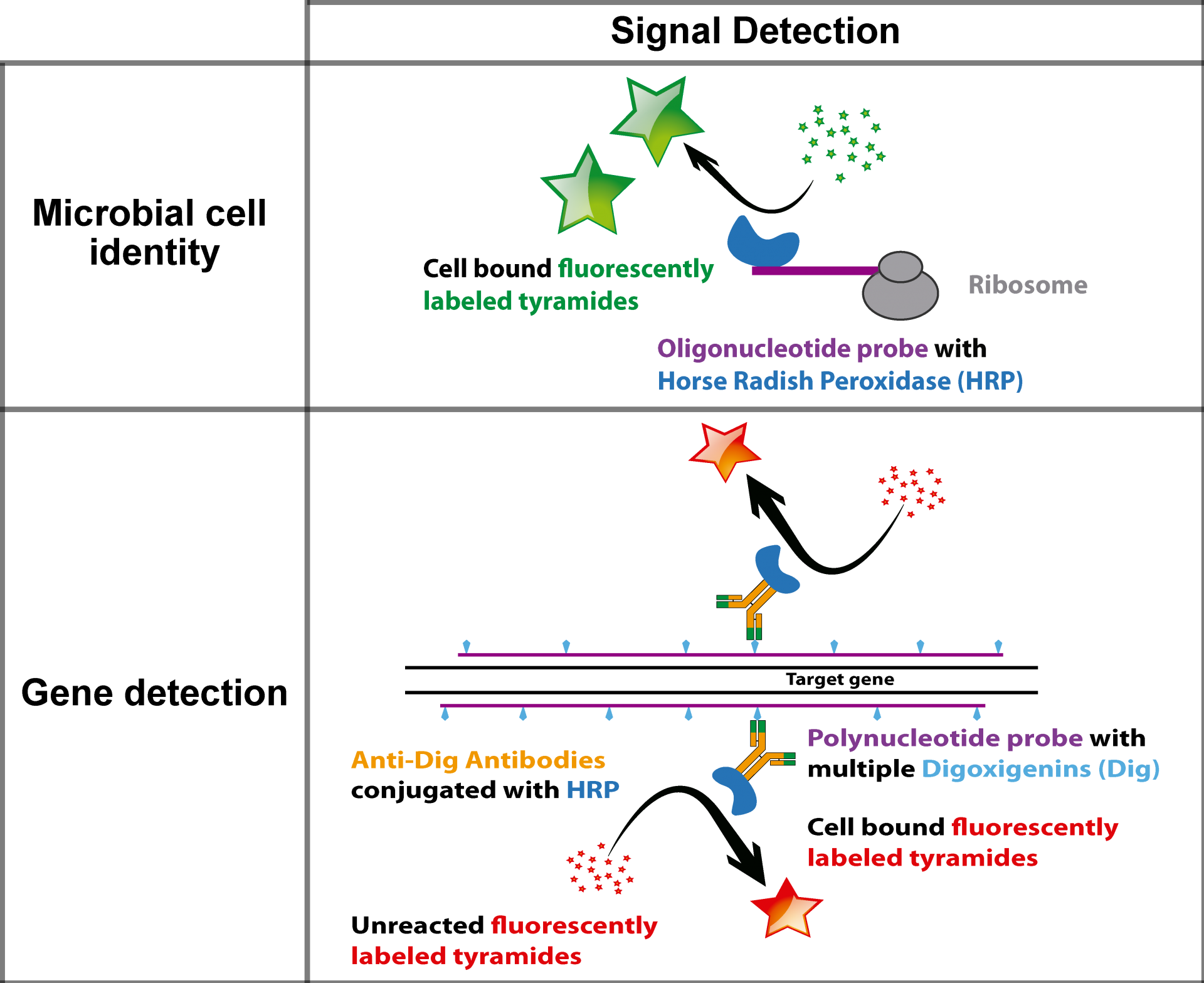
To get stronger and brighter signals of cells, we make use of an enzymatic reaction. Instead of direct labeling with a fluorescent dye, we use probes with an enzyme, a so-called peroxidase. In a second step, following the FISH hybridization, the dye molecules are added to the cells. The peroxidase enzymatically binds these dye molecules to surrounding proteinsThis way, we can also visualize microorganisms that contain particularly few rRNA sequences and which we were unable to detect using the conventional method.
GeneFISH
Detecting and Visualizing single genes in bacterial cells
GeneFISH combines the detection of specific genes and ribosomal RNA (rRNA) at the single cell level. While the presence of a certain gene is indicative of a specific functional trait, the rRNA allows for the taxonomic affiliation of the microorganism. Thus, geneFISH enables us to directly link a potential metabolic function to microbial taxonomy. We target the gene of interest with dsDNA polynucleotide probes labeled multiple digoxigenins to which horse-raddish-peroxidase labeled antibodies bind. In a second step the genes are visualized by a CARD reaction. Alternatively, polynucleotide probes are directly labeled with multiple fluorophores (direct-geneFISH). Usually, we combine geneFISH with 16S rRNA FISH to identify a group of microorganisms and study the presence of key metabolic genes.
Read more about geneFISH in this original publication: GeneFISH – an in situ technique for linking gene presence and cell identity in environmental microorganisms
Or about Direct-geneFISH: Direct-geneFISH: a simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms
Identifying phage-infected cells by phageFISH
Phages are viruses that attack bacterial and archaeal cells.The geneFISH method has been expanded to detect single phage genes. We can, thus, identify microorganisms which have been infected by a virus, as well as identifying free-floating phages.
Read more about phageFISH in this original publication: Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses.
© Max Planck Institute for Marine Microbiology/J. Barrero-Canosa