Page path:
- Press Office
- Press releases 2012
- 28.02.2012 Introns found in bacterial 16S rRNA
28.02.2012 Introns found in bacterial 16S rRNA
The characteristic that microbiologists use to determine the phylogenetic affiliation in bacteria needs to meet certain demands. It has to be conservative in its function and present in all organisms. A certain gene meets this requirement to a high degree: the gene for the smaller one of the two subunits that together form the ribosome, also known as 16S rRNA gene. For that reason it is frequently used as phylogenetic marker in analyses that researchers perform to determine the degree of relatedness in microbes.
Verena Salman from the Max Planck Institute for Marine Microbiology in Bremen has, together with her colleagues, taken a closer look at the 16S rRNA gene of a certain bacterial group, the large sulfur bacteria (Figure 1). They found out that the 16S rRNA gene of some members of this group is interrupted by long DNA inserts at up to four sites (Figure 2). However, these so called introns are not DNA inserts that encode for 16S rRNA: They either consist of non-coding DNA or they carry genes for certain enzymes by which the intron can spread into other parts of the genome.
Verena Salman from the Max Planck Institute for Marine Microbiology in Bremen has, together with her colleagues, taken a closer look at the 16S rRNA gene of a certain bacterial group, the large sulfur bacteria (Figure 1). They found out that the 16S rRNA gene of some members of this group is interrupted by long DNA inserts at up to four sites (Figure 2). However, these so called introns are not DNA inserts that encode for 16S rRNA: They either consist of non-coding DNA or they carry genes for certain enzymes by which the intron can spread into other parts of the genome.
Figure 1: The cells of the large sulfur bacteria Thiomargarita often occur in chains. In their 16S rRNA gene the scientists found four introns. At a diameter of 200 μm the cells are extremely large for bacterial cells. Image: V. Salman
“Each intron has a length of several hundred base pairs. Hence, they increase the length of the 16S rRNA gene considerably. We found 16S rRNA genes of up to 3500 base pairs length”, says Verena Salman. The common 16S rRNA gene is only 1500 base pairs long. The scientists succeeded for the first time to prove the existence of these introns in the marker gene of bacterial 16S rRNA. This is surprising, since this gene is the most sequenced gene today.
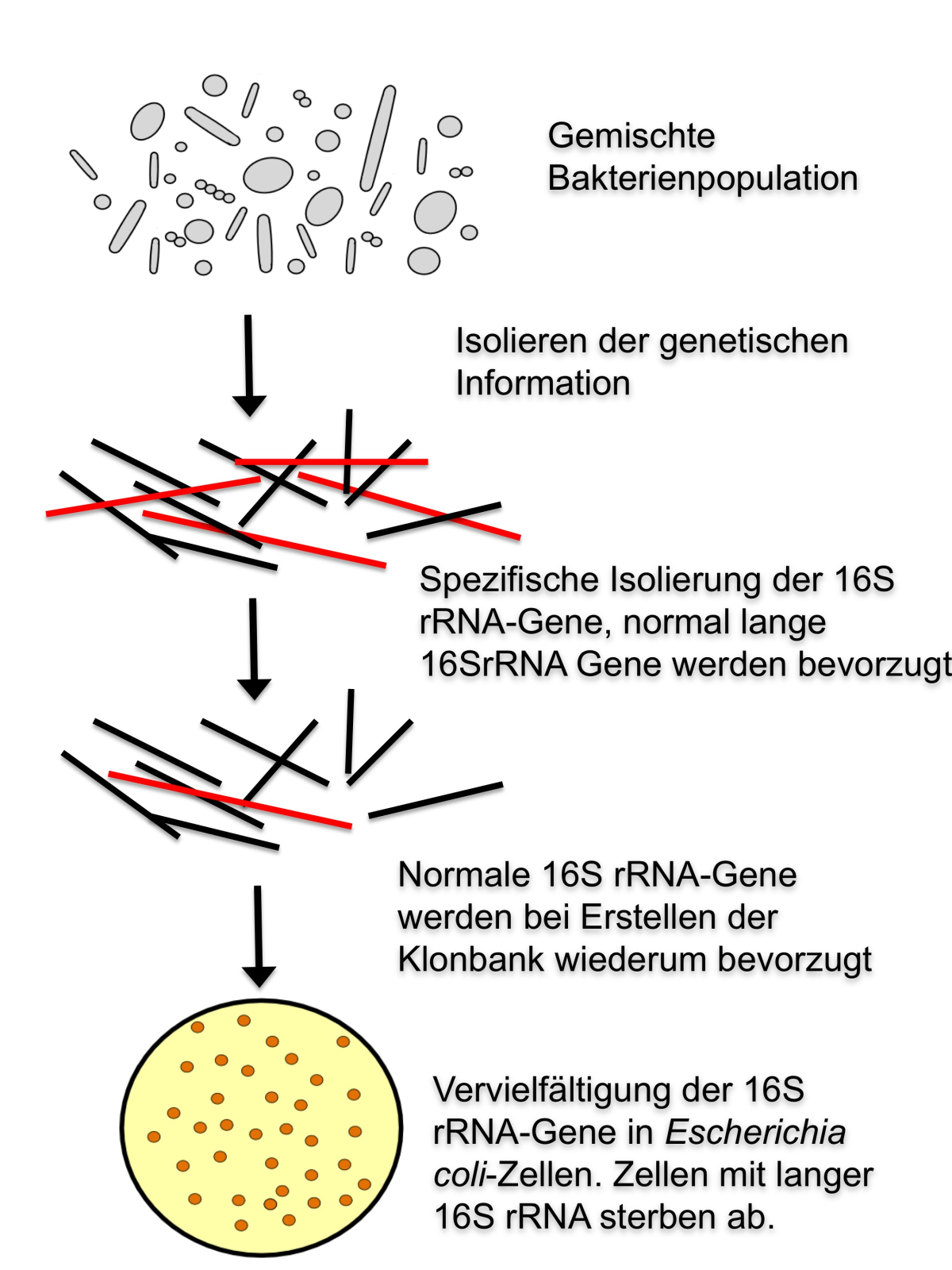
The discovery of the introns in the 16S rRNA gene has substantial implications for the microbiologists. “So far, we could only sequence short pieces of the 16S rRNA genes of the large sulfur bacteria from a mixed bacterial sample. Now we know why: Their 16S rRNA gene is longer than that of other bacteria,” explains Rudolf Amann, director of the Max Planck Institute. “Microbial cells that carry the long version of the gene cannot be identified by the classical methods, because the isolation of shorter sequences is strongly preferred through these methods.” For that reason the scientists could so far not detect the gene in its full length (Figure 3).
The discovery of the introns in the 16S rRNA gene has substantial implications for the microbiologists. “So far, we could only sequence short pieces of the 16S rRNA genes of the large sulfur bacteria from a mixed bacterial sample. Now we know why: Their 16S rRNA gene is longer than that of other bacteria,” explains Rudolf Amann, director of the Max Planck Institute. “Microbial cells that carry the long version of the gene cannot be identified by the classical methods, because the isolation of shorter sequences is strongly preferred through these methods.” For that reason the scientists could so far not detect the gene in its full length (Figure 3).
Figure 2: In the schematic view of the 16S rRNA gene the positions of the introns are marked in light grey. The introns are removed before the genetic information of the ribosome is read. Image: V. Salman/R. Dunker
Since the 16S rRNA gene plays an important role in the phylogenetic classification of bacteria and in biodiversity studies, the discovery of the scientists from Bremen has far-reaching consequences. Presumably, the introns also occur in the 16S rRNA genes of other bacterial groups. “Currently we try to find out which other microorganisms also carry the elongated 16S rRNA gene”, as Verena Salman says. “With this information we will develop a strategy that also considers the long 16S rRNA sequences, for instance in biodiversity studies”.
The bacteria do not seem to be affected by the introns in the 16S rRNA genes. Despite the long introns the cells of the large sulfur bacteria develop functional ribosomes. The introns remove themselves during the ribosome formation process (Figure 2).
The bacteria do not seem to be affected by the introns in the 16S rRNA genes. Despite the long introns the cells of the large sulfur bacteria develop functional ribosomes. The introns remove themselves during the ribosome formation process (Figure 2).
Figure 3: 16S rRNA-genes from bacteria in a mixed sample can only be detected by the standard methods if they belong to a certain length scheme. Longer 16S rRNA genes, e.g. those with inserts, remain undetected and hence are missing in biodiversity studies. The normal 16S rRNA gene is marked in black, the elongated version in red. Image: V. Salman/R. Dunker
For further information please contact
Dr. Verena Salman
Prof. Dr. Heide Schulz-Vogt
Prof. Dr. Rudolf Amann
Or the public relations office
Dr. Rita Dunker
Dr. Manfred Schlösser
Original article
Multiple self-splicing introns in the 16S rRNA genes of giant sulfur bacteria, 2012. V. Salman, R. Amann, D. A. Shub, and H. Schulz-Vogt. Proceedings of the National Academy of Science, Early Edition.
DOI: 10.1073/pnas.1120192109
Involved institutions
Max Planck Institute for Marine Microbiology, Bremen, Germany
University of Albany, State University of New York, Albany, USA
Dr. Verena Salman
Prof. Dr. Heide Schulz-Vogt
Prof. Dr. Rudolf Amann
Or the public relations office
Dr. Rita Dunker
Dr. Manfred Schlösser
Original article
Multiple self-splicing introns in the 16S rRNA genes of giant sulfur bacteria, 2012. V. Salman, R. Amann, D. A. Shub, and H. Schulz-Vogt. Proceedings of the National Academy of Science, Early Edition.
DOI: 10.1073/pnas.1120192109
Involved institutions
Max Planck Institute for Marine Microbiology, Bremen, Germany
University of Albany, State University of New York, Albany, USA