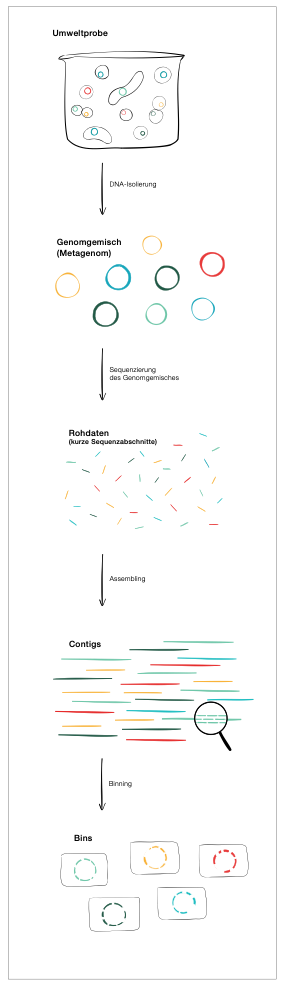Metagenomics
What is metagenomics?
Imagine a pile of beautiful books in front of you. Now imagine they have been cut into small snippets, each containing individual words and sentence fragments. You are now given the task of bringing order to this chaos. The best way to start would be to search for keywords. If the snippet says "Hamlet", it probably belongs to Shakespeare’s work of the same name. And if "new speak" or "Big Brother" comes up, you’re probably dealing with George Orwell’s 1984. "Muggles" and "Hogwarts" would clearly point to the Harry Potter series. In this way, you can dig through the pile of paper, piece by piece, and try to sort and reassemble the snippets. If several copies of the same book have been cut up differently, this makes things a bit easier. Overlapping text fragments will give you additional clues as to the correct composition of the texts.
Metagenomics isn’t all that different. Instead of books, researchers work with the genomes of micro-organisms that they cut up and reassemble. The snippets are fragments of the genome or "sequence segments". Computers can help to assemble the individual parts. There are now excellent algorithms for dealing with such a task. Some sequence segments are also already known and stored in databases. These can help to assign certain sequences to groups of organisms or functions.
With the help of metagenomics, it is possible to analyse an environmental sample (i.e. a mixture of the most diverse organisms as they live together in the environment) and find out which organisms live in this environment as well as which genes – and thus metabolic pathways, interactions, and defence strategies – predominate. This distinguishes meta-genomics from classical microbiological methods, which specifically search for individual organisms (groups) for which the genome sequences are known. Bioinformatics refers to the methods used to bring order to the metagenomic analyses and data.
Metagenomic methods also allow us to identify microorganisms – regardless of whether they can be cultured in the laboratory – and thus to study the dynamics in populations over days and even years. We can thus gain insight into the complexity and change of the communities our microscopic co-inhabitants. However, metagenomics often cannot stand alone; without classical microbiology, we would be unable to interpret the results.
How do we carry out a metagenomic analysis?
In an environmental sample, we find a mixture of the genomes of various organisms, the metagenome. In order to efficiently sequence the meta-genome of a bacterial community, for example, we must first disassemble the individual genomes into countless fragments. Sequencing provides us with numerous unordered sequence segments (depending on the technology used) ranging from 100 to hundreds of thousands of bases in length.
Despite the enormous progress in read lengths, the fragments are much shorter than the typical genomes of microorganisms. The pieces must then be assembled into larger fragments or "contigs", which are contiguous sets of overlapping pieces of DNA that come from the same genetic source – in our case, the same species of bacteria. These range in length from several kilobases to one Megabase and are thus long enough to be sorted sensibly.
We can then group the contigs in such a way that they yield more or less complete genomes of individual bacterial species. In this "binning" process, the contigs are grouped into "bins" depending on their base sequence and frequency. A bin is where pieces of the large meta-genomic puzzle that are most likely to come from a certain bacterial species are sorted. Once they are sufficiently complete, these bins are referred to as metagenome assembled genomes (MAGs).
The graphic visualizes the principle.
Metagenomics in action
The molecular ecology of algal blooms
For example, we use meta-genomics to study the degradation of algal blooms in the North Sea: every year in spring, when the days get longer and the sunlight increases, many phytoplankton species multiply rapidly – and their population grows exponentially. These algal blooms are of global importance for the carbon cycle. During photosynthesis, the small algae convert carbon dioxide into sugar compounds and oxygen. Within a few weeks, the algae run out of nutrients, and predators spread. The algae bloom then collapses. The bacteria then multiply en masse. They decompose the remaining algal biomass, and the bound carbon is largely released in the process. However, these processes are still not well understood. Which marine bacteria grow when? How do they process the algal biomass and release the carbon? What processes are behind this?
Example Projects
Simpler than expected: A microbial community with reduced diversity cleans up after algal blooms
Algae blooms regularly make for pretty, swirly satellite photos of lakes and oceans. They also make the news occasionally for poisoning fish, people and other animals. What's less frequently discussed is the outsize role they play in global carbon cycling. A recent study now reveals surprising facts about carbon flow in phytoplankton blooms. Unexpectedly few bacterial clades with a restricted set of genes are responsible for a major part of the degradation of algal sugars.
Here you can find the press release: https://www.mpi-bremen.de/en/Simpler-than-expected-A-microbial-community-with-reduced-diversity-cleans-up-after-algal-blooms.html

Lost at sea: Far off the coast, Thioglobus perditus lives off its reserve pack
SUP05 bacteria are often found in places where there is really no basis for life for them. Researchers in Bremen have now discovered that they are even quite active there – possibly with consequences for the global nitrogen cycle. The bacteria travel with a “reserve pack”. In addition, the researchers have deciphered the bacteria’s genome. The results have now been published in the journal Nature Communications.
Here you can find the press release: https://www.mpi-bremen.de/en/Lost-at-sea-Far-off-the-coast-Thioglobus-perditus-lives-off-its-reserve-pack.html
Many cooks don't spoil the broth: Manifold symbionts prepare their host for any eventuality
Deep-sea mussels, which cooperate with symbiotic bacteria for their food, harbor a surprisingly high diversity of these bacterial “cooks”: Up to 16 different bacterial strains live in the mussel's gills, each with its own abilities and strengths. Thanks to this diversity of symbiotic bacterial partners, the mussel is prepared for all eventualities. The mussel bundles up an all-round carefree package, a German-Austrian research team around Rebecca Ansorge and Nicole Dubilier from the Max-Planck-Institute for Marine Microbiology in Bremen and Jillian Petersen from the University of Vienna now reports in Nature Microbiology.
Here you can find the press release: https://www.mpi-bremen.de/en/Many-cooks-don-t-spoil-the-broth-Manifold-symbionts-prepare-their-host-for-any-eventuality..html