Page path:
- Press Office
- Press releases 2011
- 12.05.2011 New standards for genomic data
12.05.2011 New standards for genomic data
How to handle priceless sequence data
MIMARKS and MIxS - novel minimum information standards for marker genes and any (x) sequence in environmental research
An international group of nearly 100 scientists from the Genomic Standards Consortium (GSC) led by researchers from the Microbial Genomics and Bioinformatics Group at the Max Planck Institute for Marine Microbiology in Bremen, NERC Centre for Ecology and Hydrology (CEH), and University of Colorado has proposed a new standard and universal specifications that will vastly improve the ability to interpret data from marker gene studies. As of today, the majority of sequence datasets in the public repositories are lacking even the basic information about habitat parameters or the exact geographic location of the samples that have been sequenced. Now the GSC group presents the Minimum Information about a MARKer gene Sequence (MIMARKS) as the newest checklist in the Minimum Information about any (x) Sequence (MIxS) specifications in Nature Biotechnology (doi:10.1038/nbt.1823).
MIMARKS and MIxS - novel minimum information standards for marker genes and any (x) sequence in environmental research
An international group of nearly 100 scientists from the Genomic Standards Consortium (GSC) led by researchers from the Microbial Genomics and Bioinformatics Group at the Max Planck Institute for Marine Microbiology in Bremen, NERC Centre for Ecology and Hydrology (CEH), and University of Colorado has proposed a new standard and universal specifications that will vastly improve the ability to interpret data from marker gene studies. As of today, the majority of sequence datasets in the public repositories are lacking even the basic information about habitat parameters or the exact geographic location of the samples that have been sequenced. Now the GSC group presents the Minimum Information about a MARKer gene Sequence (MIMARKS) as the newest checklist in the Minimum Information about any (x) Sequence (MIxS) specifications in Nature Biotechnology (doi:10.1038/nbt.1823).
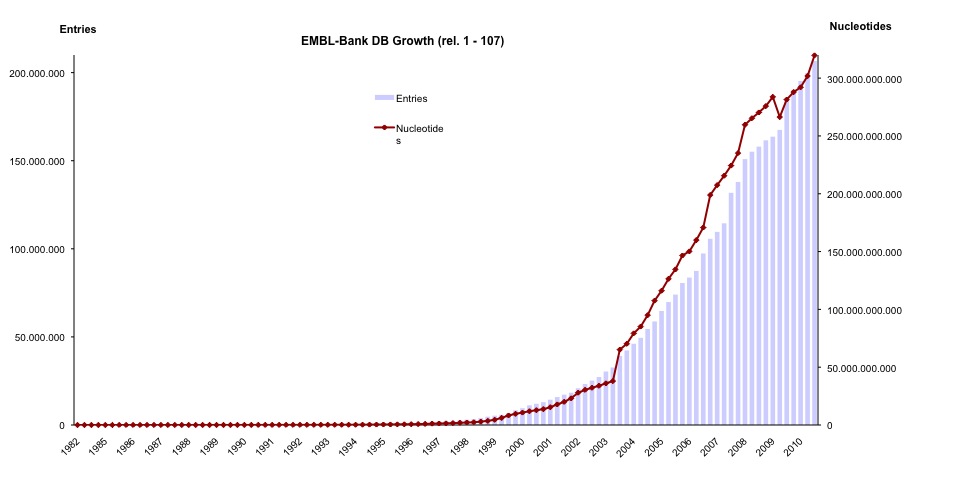
Graph showing the exponential increase of entries in the public repositories (INSDC) for the last 30 years
The pace at which sequencing projects generate data is increasing dramatically. Due to the development of new high-throughput techniques, science has entered an era of mega-sequencing projects analysing organisms in different habitats on earth, in the oceans and even inside the human body. Scientists collect samples and deposit the sequence data in public nucleic acid sequence data banks (European Nucleotide Archive, GenBank, and the DNA Databank of Japan) represented by the International Nucleotide Sequence Database Collaboration (INDSC). Unfortunately, in most cases valuable contextual (meta)data about the geographic location, the sample collection time, the habitats and other circumstances are missing. This leaves other scientists interested in these data with the time-consuming work of browsing through the literature, contacting the authors directly, or finding out that this information no longer exists.
Prof. Frank Oliver Glöckner from the Max Planck Institute and Jacobs University in Bremen says: “Sequence information without contextual (meta)data is like a new fancy tool without a manual. You can guess about its function, but without exact specifications any usage will be limited. We hope that the new Minimum information about a marker gene sequence (MIMARKS) and Minimum Information about any (x) Sequence (MIxS) specifications will greatly facilitate the ability to retrieve appropriate contextual data for marker genes and enable a new dimension of meta-analyses not otherwise feasible.”
The authors of the MIMARKS and MIxS projects are aware that this effort requires the active collaboration and participation of the scientific community in standardizing the contextual datasets in terms of content, syntax and terminology. In 2005, researchers from different fields met for the first time and founded the Genomic Standards Consortium (GSC), an open forum, in order to pave the way towards better descriptions of the genomes, metagenomes and related data (Field D et al. (2008) The Minimum Information about a Genome Sequence (MIGS) specification. Nature Biotechnology 26:541-547). After an additional two years of intensive discussions with experts from many fields, they formulated the crucial parameters of the MIMARKS checklist and MIxS specifications now online at http://gensc.org/gc_wiki/index.php/MIMARKS and http://www.gensc.org/gc_wiki/index.php/MIxS, respectively.
Encouragingly, the present effort has participation from a broad cross-section of the community, including leading scientists, large consortia, sequencing centers, and researchers at all levels of their careers, from students to distinguished professors, reinforcing the commitment of participants in the field of microbial ecology to the free and open exchange of critical research data.
“These new rules will make the lives of future researchers easier”, says Dr. Renzo Kottmann. Dr. Rob Knight of the University of Colorado points out the importance of this standard by adding: “Every investigator will benefit immensely by being able to obtain a rapid, comprehensive answer to the question Have my microbes been seen before, and, if so, where, with whom, and under which environmental conditions?”. Prof Dawn Field of CEH adds “MIMARKS builds beautifully on the existing GSC minimum information standards MIGS and MIMS, rounding out a concise family of standards for describing genomes, metagenomes and now gene marker sequences. This publication marks a tremendous effort on the part of the community and expectations for wide-spread adoption are high.”
Manfred Schlösser
For further information please contact
Prof. Frank Oliver Glöckner and Pelin Yilmaz
or the press officer Manfred Schlösser
Original articles:
Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications
Pelin Yilmaz, Renzo Kottmann, Dawn Field, Rob Knight, Frank Oliver Glöckner and around 100 other scientists. NATURE Biotechnology 2011. (doi:10.1038/nbt.1823)
and
The genomic standards consortium: bringing standards to life for microbial ecology
Pelin Yilmaz, Jack A Gilbert, Rob Knight, Linda Amaral-Zettler, Ilene Karsch-Mizrachi, Guy Cochrane, Yasukazu Nakamura, Susanna-Assunta Sansone, Frank Oliver Glöckner and Dawn Field doi:10.1038/ismej.2011.39, http://www.nature.com/ismej/journal/vaop/ncurrent/full/ismej201139a.html
Prof. Frank Oliver Glöckner from the Max Planck Institute and Jacobs University in Bremen says: “Sequence information without contextual (meta)data is like a new fancy tool without a manual. You can guess about its function, but without exact specifications any usage will be limited. We hope that the new Minimum information about a marker gene sequence (MIMARKS) and Minimum Information about any (x) Sequence (MIxS) specifications will greatly facilitate the ability to retrieve appropriate contextual data for marker genes and enable a new dimension of meta-analyses not otherwise feasible.”
The authors of the MIMARKS and MIxS projects are aware that this effort requires the active collaboration and participation of the scientific community in standardizing the contextual datasets in terms of content, syntax and terminology. In 2005, researchers from different fields met for the first time and founded the Genomic Standards Consortium (GSC), an open forum, in order to pave the way towards better descriptions of the genomes, metagenomes and related data (Field D et al. (2008) The Minimum Information about a Genome Sequence (MIGS) specification. Nature Biotechnology 26:541-547). After an additional two years of intensive discussions with experts from many fields, they formulated the crucial parameters of the MIMARKS checklist and MIxS specifications now online at http://gensc.org/gc_wiki/index.php/MIMARKS and http://www.gensc.org/gc_wiki/index.php/MIxS, respectively.
Encouragingly, the present effort has participation from a broad cross-section of the community, including leading scientists, large consortia, sequencing centers, and researchers at all levels of their careers, from students to distinguished professors, reinforcing the commitment of participants in the field of microbial ecology to the free and open exchange of critical research data.
“These new rules will make the lives of future researchers easier”, says Dr. Renzo Kottmann. Dr. Rob Knight of the University of Colorado points out the importance of this standard by adding: “Every investigator will benefit immensely by being able to obtain a rapid, comprehensive answer to the question Have my microbes been seen before, and, if so, where, with whom, and under which environmental conditions?”. Prof Dawn Field of CEH adds “MIMARKS builds beautifully on the existing GSC minimum information standards MIGS and MIMS, rounding out a concise family of standards for describing genomes, metagenomes and now gene marker sequences. This publication marks a tremendous effort on the part of the community and expectations for wide-spread adoption are high.”
Manfred Schlösser
For further information please contact
Prof. Frank Oliver Glöckner and Pelin Yilmaz
or the press officer Manfred Schlösser
Original articles:
Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications
Pelin Yilmaz, Renzo Kottmann, Dawn Field, Rob Knight, Frank Oliver Glöckner and around 100 other scientists. NATURE Biotechnology 2011. (doi:10.1038/nbt.1823)
and
The genomic standards consortium: bringing standards to life for microbial ecology
Pelin Yilmaz, Jack A Gilbert, Rob Knight, Linda Amaral-Zettler, Ilene Karsch-Mizrachi, Guy Cochrane, Yasukazu Nakamura, Susanna-Assunta Sansone, Frank Oliver Glöckner and Dawn Field doi:10.1038/ismej.2011.39, http://www.nature.com/ismej/journal/vaop/ncurrent/full/ismej201139a.html