- Press Office
- Press releases
- Structures of methane-activating enzyme in elusive archaea
Structures of methane-activating enzyme in elusive archaea
MCR: an enzyme at the core of the planetary methane cycle
Methane is a very potent greenhouse gas – over 25 times more effective than carbon dioxide. Thus, every molecule that is kept from entering the atmosphere makes a significant difference. The enzyme MCR can determine whether methane is released into the air or recycled into carbon dioxide. That is because MCR catalyzes the final step of methane production in methanogens and the first step in anaerobic methane oxidation — that is, methane consumption— in ANME.
While MCR in methane-producing archaea has been studied for decades, much less is known about how it functions in ANME. These microorganisms remain difficult to study because they cannot be cultivated in pure cultures in the laboratory. Until now, only one MCR structure from ANME had been solved, from a microbial mat collected in the Black Sea.
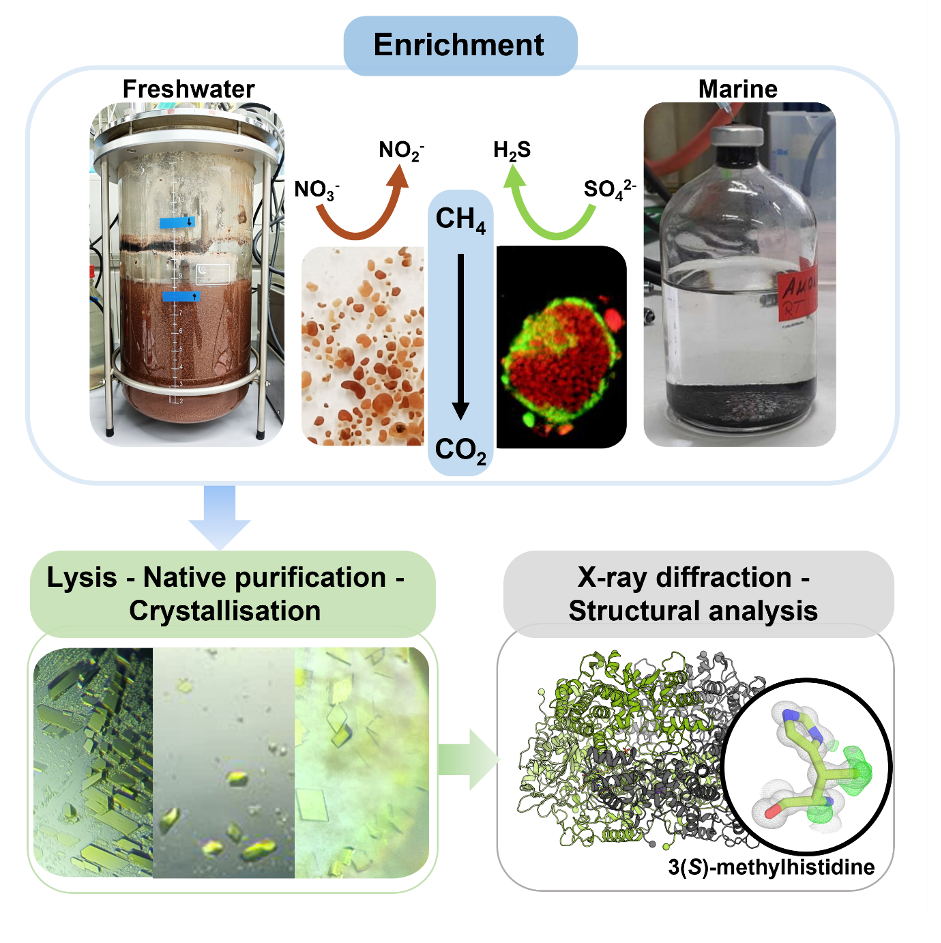
From mud volcanoes to bioreactors: enriching the uncultivable
“Since methane-consuming and methane-producing archaea (i.e., ANME and methanogens) both rely on MCRs, we wondered whether they function in the same way in both directions of the methane cycle”, says project leader Tristan Wagner from the Max Planck Institute for Marine Microbiology in Bremen (Germany). “To understand the differences between the MCR from ANME and methanogens, we needed a picture of the enzyme.” Therefore, the researchers from Bremen joined forces with partners across Europe to structurally characterize ANME´s enzyme. Colleagues at Radboud University (Netherlands) provided microbial enrichments from two distinct freshwater samples with ANME-2d archaea, which oxidize methane using nitrate, and Gunter Wegener from the Max Planck Institute in Bremen provided marine samples of ANME-2c, obtained from deep-sea sediments at the Amon Mud Volcano in the Mediterranean Sea, which form consortia with sulfate-reducing bacteria.
In these enrichments, researchers from Radboud University and the National Museum of Natural Sciences of Madrid (Spain) analyzed the microbial communities and provided genomic information revealing that MCRs were present. Wagner and his team then processed the biomass coming from the enrichments to purify and crystallize the MCRs.
Similar enzymes, unusual modifications
Unexpectedly, the yellow crystals that appeared were of outstanding quality, providing X-ray diffraction at a level that has never been reached before. The subsequent analysis revealed that MCRs from both freshwater and marine ANME were highly similar to those found in well-known methane-producing archaea, despite their metabolic differences. “This supports the idea that methane production and methane oxidation, two chemically opposite reactions, are both catalyzed by the same enzyme, just operating in reverse”, Wagner stresses. Further biophysical experiments on crystals performed by specialists from the European Synchrotron Radiation Facility and from the Institut de Biology Structurale in Grenoble (France) confirmed that the MCR from ANME should share the same reaction sequences as proposed for the methanogenic one.
The researchers discovered that the methane-processing enzyme (MCR) has a record seven chemical modifications per active site. One of these tweaks — a never-before-seen modification called 3(S)-methylhistidine, confirmed by an expert in mass spectrometry from the Max Planck Institute for Terrestrial Microbiology in Marburg (Germany)—was found near the enzyme’s key nickel cofactor. This rare modification might help the archaea break down methane more efficiently, even though its exact role is still unknown.
Learning from ANMEs is key for climate research
This study reinforces the importance of understanding enzymes in their natural context. With three ANME-derived MCR structures now available, including the only known enzyme bearing a 3(S)-methylhistidine, scientists are better equipped to explore how nature controls methane flux at the molecular level in oxygen-free environments.
“By identifying which cofactors are used and which post-translational modifications are present, this research provides valuable guidance for designing engineered systems that mimic the natural microbial methane conversion”, Wagner concludes. “Such research efforts could one day contribute to reducing methane emissions and support global efforts toward a future with zero-carbon emissions.”
Original publication
Marie-C. Müller, Martijn Wissink, Priyadarshini Mukherjee, Nicole Von Possel, Rafael Laso-Pérez, Sylvain Engilberge, Philippe Carpentier, Jörg Kahnt, Gunter Wegener, Cornelia U. Welte, Tristan Wagner (2025): Atomic resolution structures of the methane-activating enzyme in anaerobic methanotrophy reveal extensive post-translational modifications. Nature Communications, DOI: 10.1038/s41467-025-63387-1
Participating institutions
- Max-Planck-Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany
- Department of Microbiology, Radboud Institute for Biological and Environmental Sciences (RIBES), Radboud University, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands
- Biogeochemistry and Microbial Ecology Department, Museo Nacional de Ciencias Naturales (MNCN-CSIC), C. de José Gutiérrez Abascal, 2, 28006, Madrid, Spain
- Univ. Grenoble Alpes, CEA, CNRS, Institut de Biologie Structurale, 71 avenue des Martyrs, 38043 Grenoble, France
- European Synchrotron Radiation Facility, 71 avenue des Martyrs, 38043 Grenoble, France
- Univ. Grenoble Alpes, CEA, CNRS, IRIG-LCBM UMR 5249, 17 avenue des Martyrs, 38054 Grenoble, France
- Max Planck Institute for Terrestrial Microbiology, Karl-von-Frisch-Strasse 10, 35043 Marburg, Germany
Please direct your queries to:
Head of Group
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany

Head of Press & Communications
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
1345 |
|
Phone: |

