- Press Office
- Press releases 2020
- Analysis of unknown microbes is getting easier
Analysis of unknown microbes is getting easier
Using a combination of several methods, the scientists from Bremen have managed to describe the functional capacities of less common organisms in a marine microbial community using the metagenome. The metagenome is the sum of all genes existing in a microbial community. In this large amount of genetic information, the DNA of the rare microorganisms was successfully recognized and subsequently analyzed. This is important because many microbial communities are dominated by one or two species. Yet, many other organisms are also part of the community and play an important role, even though they are less numerous. Until now, it was difficult to identify their function.
A specific case concerned bacteria that were found after algae blooms off the island of Helgoland in the North Sea. “We repeatedly came across an unknown species in our samples and wanted to learn more about their functional capacities and their role in the environment,“ says Anissa Grieb, researcher at the Max Planck Institute for Marine Microbiology and first author of the study. But as these bacteria could neither be grown in the lab nor be found via the metagenome using standard methods, the team with Grieb started to look for other solutions.
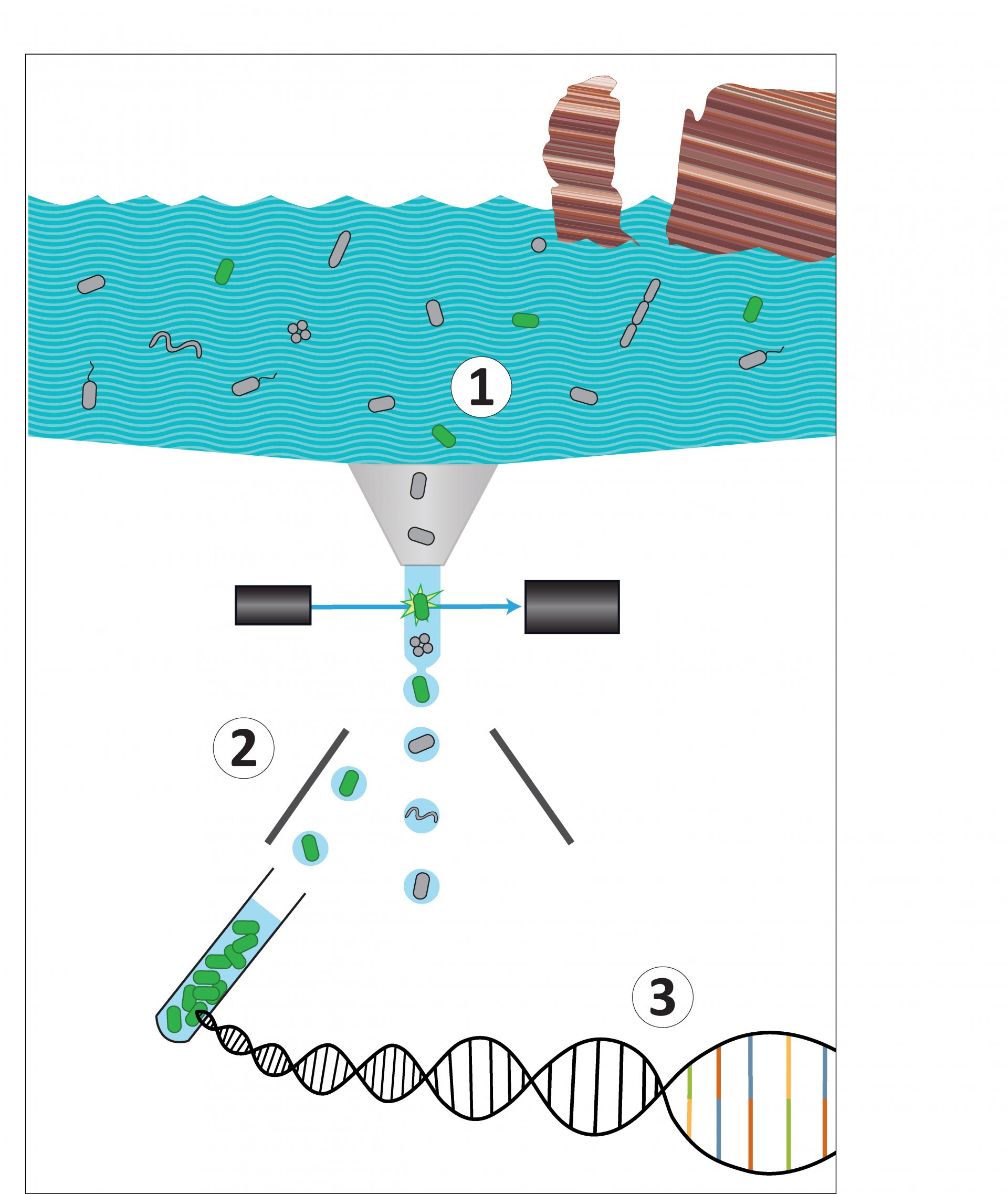
The good into the pot
The idea: „As we have a flow cytometer with a cell sorting system in our research group, we had the idea to select the cells before sequencing the DNA“, says Grieb. That way, the diversity is reduced and the rare species can no longer hide. Thus, the scientists used the FISH-method to label DNA sections of the microbes with fluorescent dyes to detect and sort the bacteria they were interested in. “However, we faced two challenges: Firstly, a very bright FISH signal was needed for the detection and sorting of targeted cells using flow cytometry,” says Grieb. “Secondly, sufficient unimpaired DNA material was required for high quality genome sequencing.”
The team around Grieb, together with researchers from the Joint Genome Institute, tested how this could work. At the end, they succeeded by using a recently developed version of the FISH-method, the hybridization chain reaction (HCR)-FISH. First, they optimized this procedure with pure cultures in the laboratory and afterwards used it on the environmental samples, which had been taken off the coast of Helgoland.
Microbes become visible
“As a result, we are able to isolate the genome of rare microorganisms and analyze it afterwards with standard-methods of gene analysis,” says Bernhard Fuchs, leader of the flow cytometry research group at the Max Planck Institute for Marine Microbiology. “Specific groups or species can now be genomically interrogated, opening a window into the biosphere of rare microorganisms, even of highly complex samples.” This new method enabled the researchers to clarify the identity of a less common genus, whose DNA repeatedly appeared in water samples from the North Sea off Helgoland. In other words: The game of hide and seek is over.
Original publication
Anissa Grieb, Robert M. Bowers, Monike Oggerin, Danielle Goudeau, Janey Lee, Rex R. Malmstrom, Tanja Woyke and Bernhard M. Fuchs: A pipeline for targeted metagenomics of environmental bacteria. Microbiome 8, February 2020.
Behind the paper: Finding the needle in a haystack via targeted genomics
Participating institutions
- Max Planck Institute for Marine Microbiology, Bremen, Germany
- DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, USA
Please direct your queries to:
Group Leader
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
2222 |
|
Phone: |


