- Departments
- Department of Molecular Ecology
- Coastal Sediments
Coastal Sediments
Project Leader
Project leader
Department of Molecular Ecology
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
2222 |
|
Phone: |

Our research
Our aim is to understand carbon cycling in coastal sandy sediments with a focus on heterotrophic bacterial communities that live in surface layers. Research questions address, for example, if seasonality in primary production is reflected in benthic bacterial community composition, the identification of main food sources for benthic heterotrohic bacteria (algae-derived or animal-derived carbon) or different degradation modes for polysaccharides by specific benthic taxa (selfish bacteria versus external hydrolyzers versus scavenging bacteria).
In the oceans, about one-fifth of primary production takes place at continental shelves (Jahnke, 2010), emphasizing the importance of these ecosystems for global carbon cycling. The first interaction of water column-derived organic matter with benthic microbial communities takes place in surface sediments which are acting as biological filters catalyzing central steps of elemental cycling.
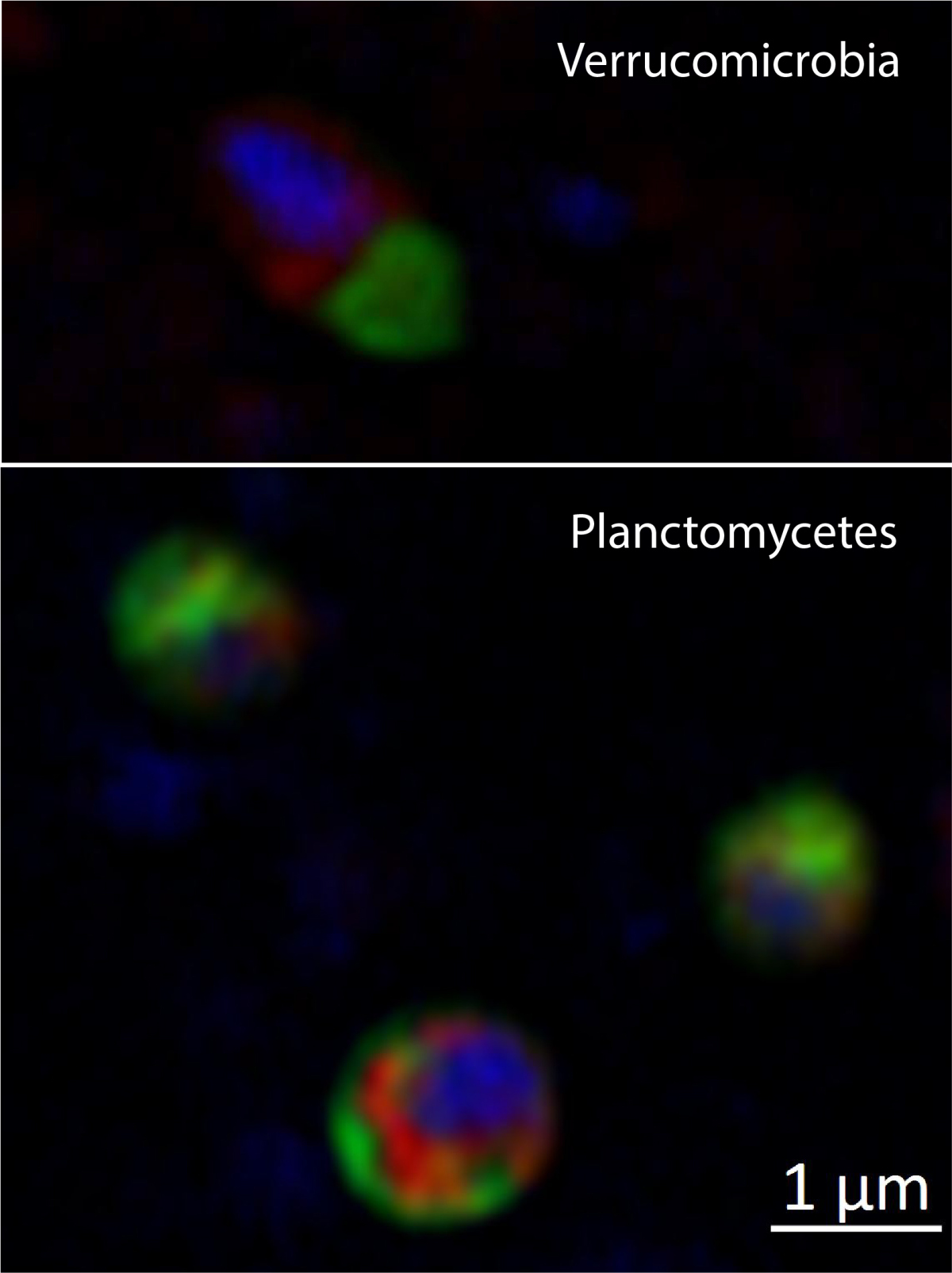
Helgoland Roads (North Sea, German Bight) and Isfjorden (Svalbard, Arctic Ocean) are our main study sites. Bacterial communities in surface sediments were richer, more even and significantly different from communities in bottom waters (Probandt et al. 2017; Miksch et al., subm.). Planctomycetes, Verrucomicrobia and Actinobacteria are suggested as key bacteria for degradation of high molecular weight compounds and recalcitrant material that entered surface sediments from the water column.
In October 2021, we received a donation of 10,000 euros from the Andreas Rühl Foundation. The donation supports us in continuing the research described here.
We are very happy about this generous support!
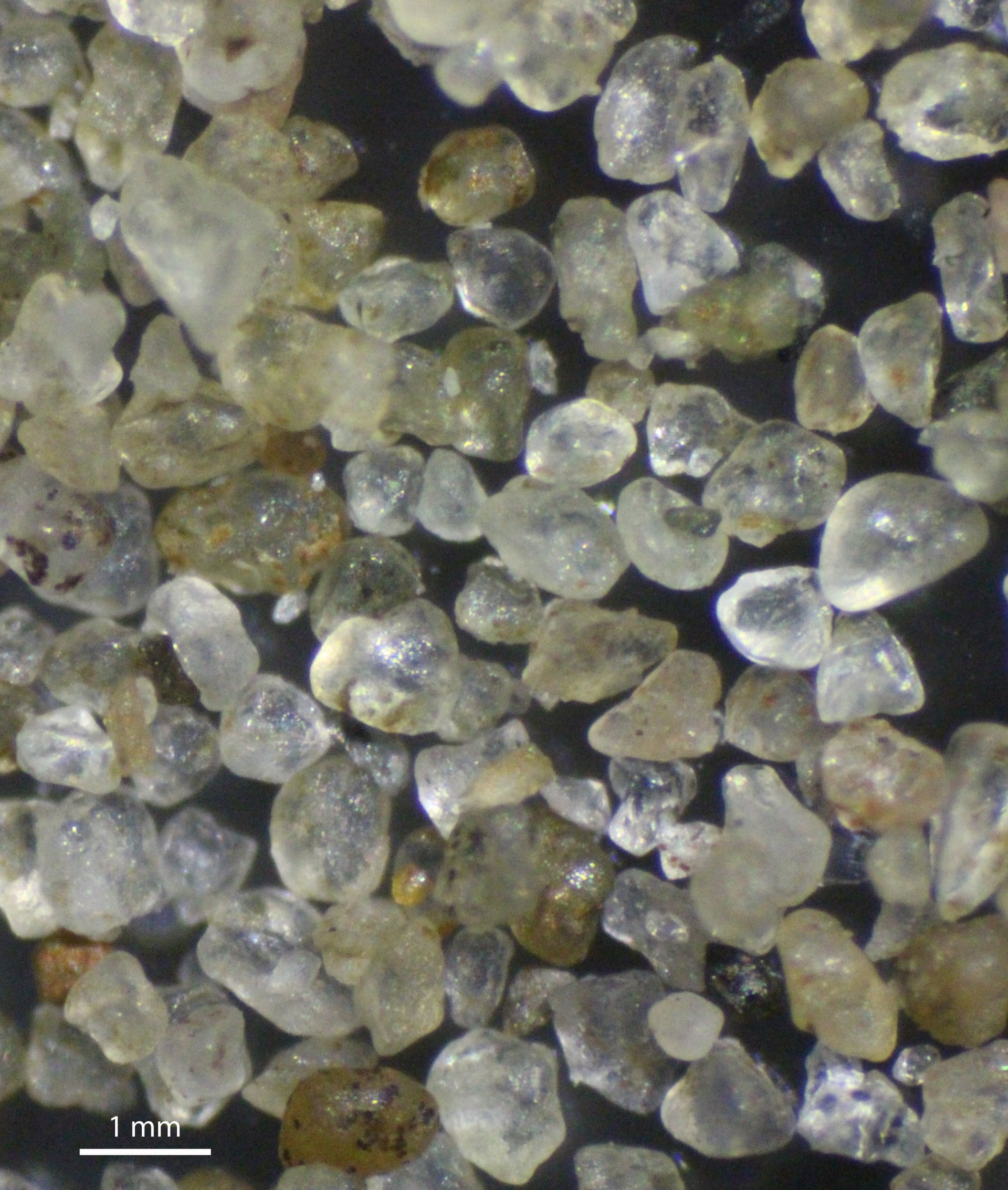
Life on sand grains
Marine sediments constitute the natural habitat for estimated 1.7 x 1028 bacteria and archaea (Whitman et al., 1998). In surface sediments, cell abundances are 108 to 109 per gram, and even for the subsurface seabed, more than 105 cells per gram have been reported. The benthic microbial community is highly diverse comprising several thousand different species (Probandt et al., 2017; Miksch et al., 2021).
The vast majority of the benthic microbial community lives attached to sand grains. Resuspension of sediment grains exposes the microbial community to mechanical shearing stress and constantly changing environmental conditions.
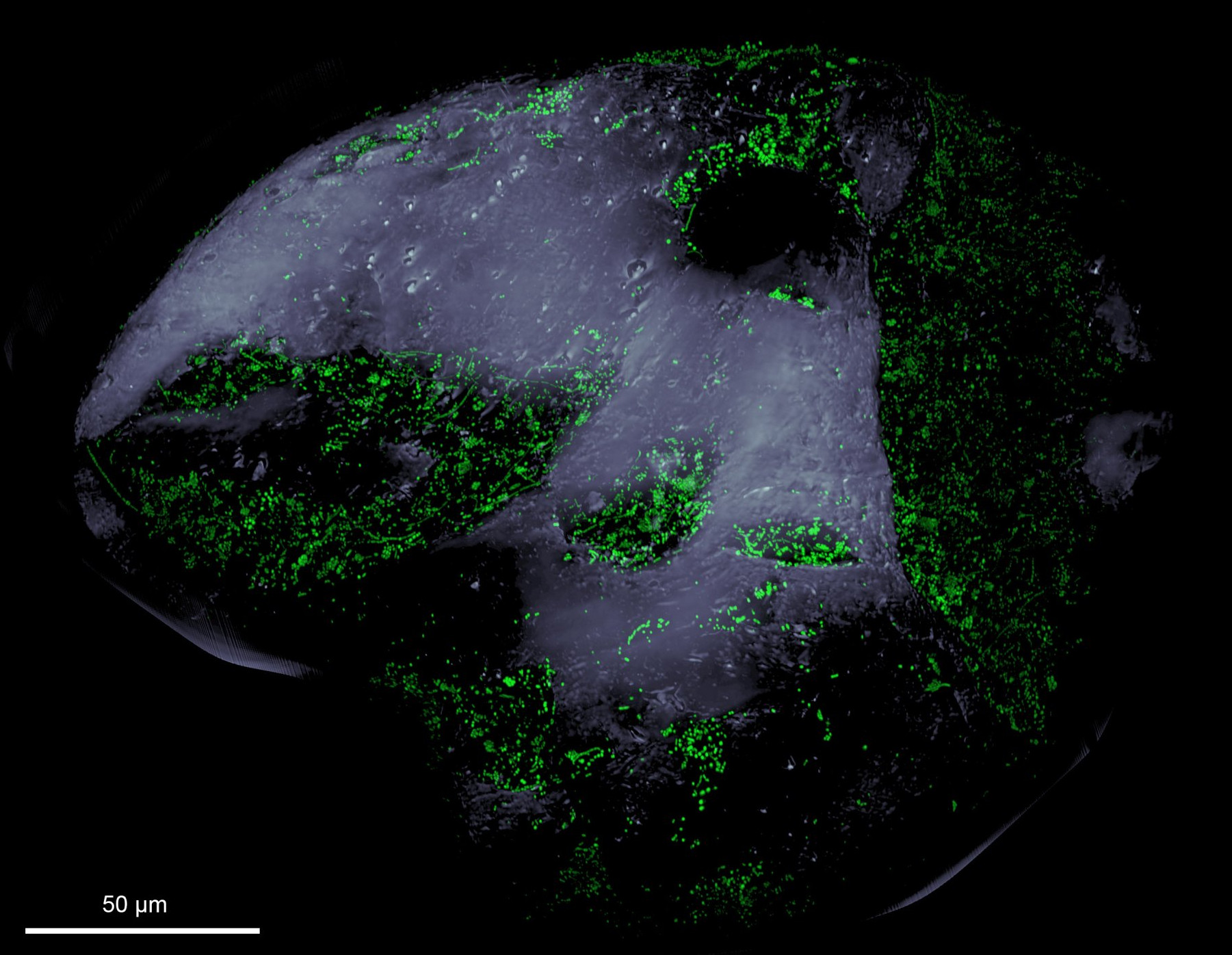
In this project we took the step from bulk sediments to single sand grains by taking a direct look at single sand grains to study the microbes in their micro-habitat (Probandt et al., 2018). Adapted protocols for hybridization or PCR of single sand grains without prior sonication or DNA extraction allows us to study the microbial community composition and structure in situ.
Each sand grain harbored a total of 104–105 cells consisting of a highly diverse bacterial community with several thousand species. Although bacterial communities differed between sand grains, a core community accounting for >50% of all species was present on each sand grain (Probandt et al. 2018). Colonization was patchy, with exposed areas largely devoid of any epi-growth and protected areas more densely populated.
As a follow-up, we started colonization experiments with sterile sand using different starting communities in vitro and in vivo (S. Silva) with the aim to understand the ecological succession of colonization.
Sediment microbes occupy different niches within the sediment
Microbes living in the pore space of the sediments make up about 3% of total cells. Other microbes are loosely attached or firmly attached to the grains' surface and comprise 8-13% and 84-89% of the total microbial community, respectively (Moncada et al., submitted). The degree of cell attachment to sediment grains is not stochastic but influenced by the surrounding microenvironment, taxa-dependent factors, and different survival strategies. Thus, these cell fractions have likely distinct roles and contributions to organic matter remineralization within surface sediments. Porewater and loosely attached cells are enriched in potential high-molecular-weight organic matter utilizing taxa that are specialized in rapid aerobic remineralization, while firmly attached cells are able to utilize not just fresh organic matter but also hydrolysis and fermentation products. Since oxygen might also become limited in local depressions, a part of this fraction must be capable of anaerobic respiration (Moncada et al., submitted).
Polysaccharide degradation
Polysaccharides are major constituents of macroalgae and phytoplankton biomass. They make up a large fraction of the organic matter produced and degraded in the oceans. Yet, little is known about identity, organization and expression of genes responsible for benthic polysaccharide degradation.
Benthic bacterial communities in sandy surface sediments are seasonally stable and do not respond to changes in primary production/substrates. Therefore, we study expression profiles of carbohydrate-active enzymes (CAZymes), in particular of glycoside hydrolases, in surface sediments from Svalbard. Our data show that the majority of the benthic bacterial community is present and active in two contrasting seasons despite the strong seasonality in polar regions (Miksch et al., 2024). Some seasonality, however, can be found, such as degradation of beta-glucans by Bacteroidota and Gammaproteobacteria in spring. Similar to what occurs in the water column laminarin degradation is a major process in sediments during spring, while utilization of alpha-glucans, in particular glycogen, occurs throughout the year. Expression of genes for degradation of other constantly available substrates such as mucin and chitin suggests that the continuous utilization of less labile, permanently available substrates stabilizes benthic bacterial communities.
Current studies aim at the autecology of taxa degrading often complex, permanently available substrates, for example by enrichment and isolation of pure cultures using mucin and glycogen (in cooperation with J. Harder).
In cooperation with the MARUM MPG Bridge Group Marine Glycobiology (J.-H. Hehemann) we are analyzing sugars available in sediments. Microbial communities in sandy sediments digest and transform labile parts of photosynthesis-derived particulate organic matter and likely release more stable, glucose-depleted residual glycans of unknown structures, quantities and residence times into the ocean thus modulating the glycan composition of marine coastal waters (Miksch et al., 2024).
In surface ocean waters, a relatively large fraction of up to one-fourth of bacteria is capable of polysaccharide degradation by using the selfish uptake mode (Reintjes et al., 2017). In contrast, high external hydrolysis rates and a small but consistently-detected fraction of selfish bacteria characterized Svalbard surface sediments (Knittel et al., submitted). The bulk of glycan utilization seems to be catalyzed by a synergistic community-based mechanism relying on the sharing of enzymatic capabilities and the scavenging of public goods.
PhD student
Department of Molecular Ecology
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
2224 |
|
Phone: |

Former members of project
Dr. Sebastian Miksch
Dr. David Probandt
Jannika Moye

