- Press Office
- Press releases 2019
- Fuel cells in bacteria
Fuel cells in bacteria
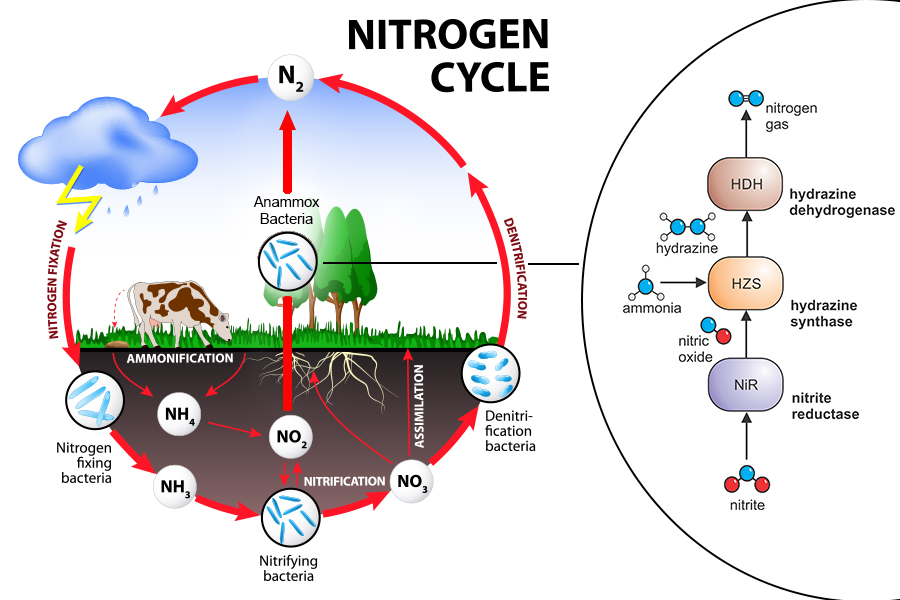
The biogeochemical nitrogen cycle
Nitrogen, in the form of nitrogen gas (N2), makes up about 80 percent of our atmosphere. All living organisms require nitrogen, because it is part of most of their essential molecules. However, they cannot use atmospheric nitrogen directly but require it in a different chemical form. A number of bacteria perform such conversions and contribute to the biochemical nitrogen cycle by producing more reactive forms of nitrogen.
Anammox bacteria - a shortcut through the middle of the nitrogen cycle
In the 1990s, scientists discovered a bacterial process called anaerobic ammonium oxidation (anammox). “We now believe this process is responsible for 30 to 70 percent of the yearly nitrogen removal from the oceans”, explains Thomas Barends, group leader at the Max Planck Institute (MPI) for Medical Research in Heidelberg. During this process, bacteria convert nitrite and ammonium into dinitrogen gas (N2) and water, while generating energy for the cell. The molecule hydrazine is produced in an intermediate step. Hydrazine is a common component of rocket fuel, but its use by bacteria as a metabolic fuel is rather exotic - and surprising in living organisms because of its high toxicity. “Production and oxidation of hydrazine is unique to anammox bacteria,” says Boran Kartal from the Max Planck Institute for Marine Microbiology in Bremen. He explains, “Earlier, we described the catalytic and spectroscopic properties of hydrazine dehydrogenase. Now our colleagues at the MPI for Medical Research in Heidelberg solved how anammox bacteria optimized the mechanism of hydrazine oxidation by using heme groups as a wiring system.”
Previously the research group and their collaborators have described the structures of the enzymes hydrazine synthase and hydroxylamine oxidoreductase. The researchers now further unravel the anammox puzzle by describing the crystal structure of hydrazine dehydrogenase, the enzyme involved in the conversion of toxic hydrazine to harmless dinitrogen gas
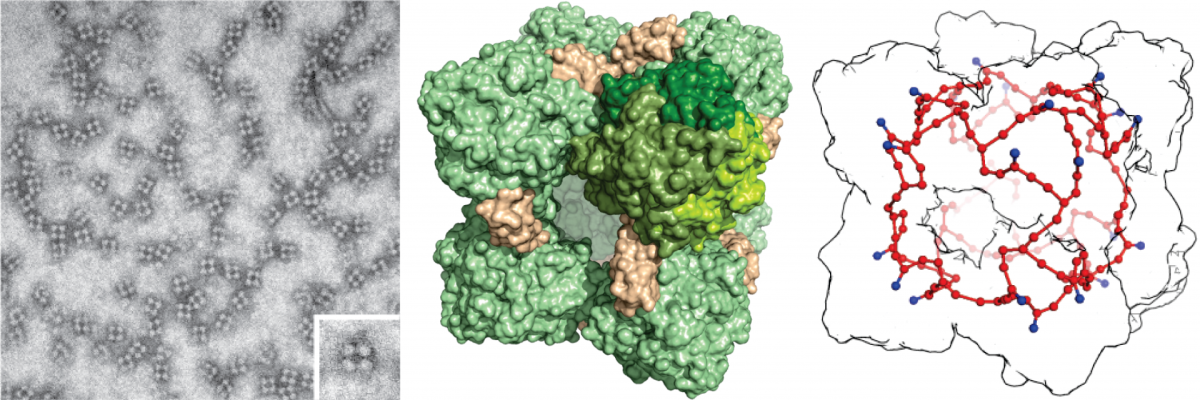
From toxic rocket fuel to harmless nitrogen – the hydrazine dehydrogenase (HDH) complex
“One could compare the HDH complex to a fuel cell with electrical outlets that only fit certain types of plugs”, says Barends, describing the structure and mechanism of HDH. The ‘fuel’ hydrazine enters the protein complex through a channel on the outside. The enzyme then catalyzes the conversion of hydrazine into nitrogen gas by an unprecedentedly large network of 192 heme groups. Then the electrons are carried to other parts of the bacterium, like the transfer of current to electrical consumers. These consumers then generate the cell’s energy.
Animation: Structure of the HDH complex. The complex has the shape, of a cube, with entrance on the side. The side length of the cube is approximately 15 nanometers. Inside the protein complex (white) is the network of heme groups (red). © Thomas Barends, Max Planck Institute for Medical Research
Closing the gap
“We are now working on finding the protein that takes up the electrons stored in the heme network”, says Mohd Akram, PostDoc in the Barends group and first author of the paper. From the structure they observed they expect that only small proteins can enter the complex, take up the electrons in a hollow space inside and leave again. Selecting which proteins can access the electrons may help ensure the electrons are brought to the right place to be used for energy generation in the cell.
Original publication:
Previous research
Participating institutions:
- Max Planck Institute for Medical Research, Heidelberg, Germany
- Max Planck Institute for Biophysics, Frankfurt am Main, Germany
- Radboud University, Nijmegen, The Netherlands
- Max Planck Institute for Marine Microbiology, Bremen, Germany
Please direct your queries to:
Group leader
Microbial Physiology Research Group
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
3126 |
|
Phone: |

Group Leader
Structural Biology of Elemental Cycles
Dr. Thomas Barends
Max Planck Institut for Medical Research
Jahnstraße 29
69120 Heidelberg
phone: +49 6221 486-508