- Departments
- Department of Molecular Ecology
- Molecular Ecology
- Automated Microscopy and Cell Counting
Automated Microscopy and Cell Counting
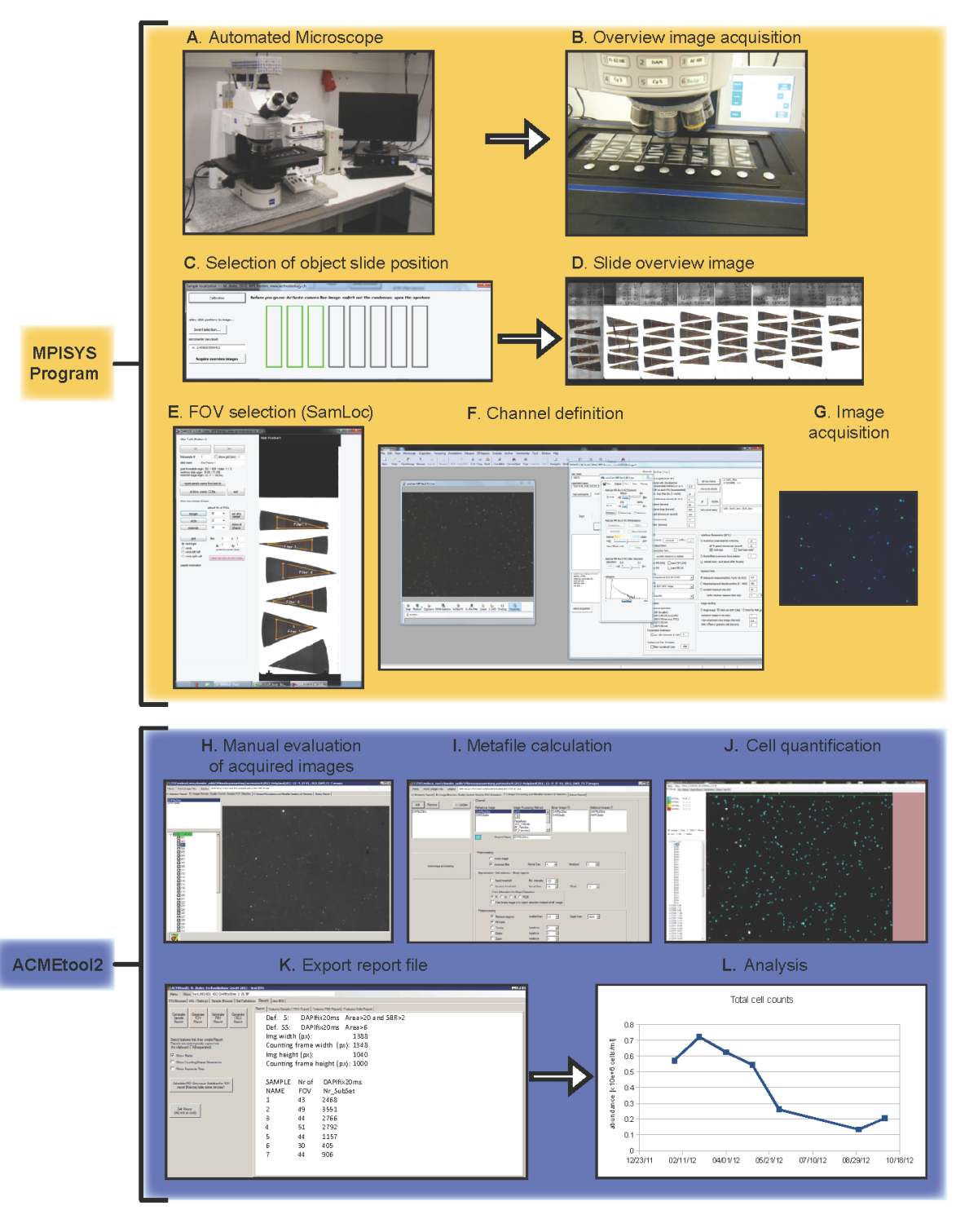
We developed an image acquisition and evaluation system for the automated microscopic evaluation of comparatively simple samples, i.e. the counting of bacterial cells in suspension that are filtered and immobilized on membrane filters. For this we have developed a robust computerized focussing system that combines a proprietary autofocus with custom texture parameters, feedback loops for image contrast maximation, and decisions on image content during acquisition.
The system is capable of excluding inappropriate or badly focussed microscopic fields prior to evaluation without operator interaction with high reliability (>95%), and to evaluate >50 FISH stained samples per day at a total of 2000-3000 counted cells per sample. It consists of two modules:
The first module MPISYS (yellow) allows for the automated scan of the specimen (in this case plankton samples on filter pieces mounted on glass slides) and the acquisition of image series from the sample.
The second module ACMEtool (blue) enumerates cells based on the images taken by MPISYS. A manual inspection of the individual images is possible to ensure a high-quality enumeration of all samples.
Instruments
- Zeiss Axioimager.Z2m
Fully motorized and software controlled microscope used for automated high-throughput microbial cell enumeration, and tile scan images of large regions or full slides.
Configuration
- PI piezo driven scanning stage for 8 microscopic glass slides.
- Microscopic image acquisition via Zeiss AxioCam 702 mono or MRm camera.
- LED illumination system (Zeiss Colibri 7) for fast switching between fluorescent channels (385, 469, 555 or 590, and 631 nm) in combination with a multiband filter set.
- Slide overview image acquisition using 1x objective lens.
The system is controlled with the 'MPISYS program' using the Zeiss AxioVision VBA software package and software tools and methods of Zeder et al., 2006 - 2011, and Bennke et al., 2016.

- Zeiss Axioplan 2
High-throughput automatic microbial cell enumeration system for shipboard analyses.
Configuration
- Märzhäuser stage EK 14 mot holding a maximum of 2 microscopic glass slides.
- Microscopic image acquisition via Zeiss AxioCam 702 mono.
- Slide overview image acquisition via Logitech 905 C webcam.
- Multi channel LED illumination system Zeiss Colibri 7 (385, 469, 555 or 590, and 631 nm).
The system is controlled with the 'MPISYS program' using the Zeiss AxioVision VBA software package and software tools and methods of Zeder et al., 2006 - 2011, and Bennke et al., 2016.
It has been successfully used during the UltraPac cruise in 2015/16 (SO 245), the Arctic cruise in 2016 (MSM 56), and during the Atlantic Meridional Transect cruise in 2012 (AMT 22).

Software Tools
- Automated Cell Counting (ACMEtool)
Automated cell counting on microscopic images is done by using the software ACMEtool.
ACMEtool (Automated Cell Measuring and Enumeration Tool) has been developed by M. Zeder (technobiology).
It is a program that allows for image analysis using multiple fluorescence channels. It was developed to analyse aquatic bacterial cells concentrated on filter membranes and stained usually with DAPI and FISH. It can also be used for simple cell counting, or more complex experiments, such as microautoradiograpy combined with FISH or geneFISH. It can yield cell shape, signal intensities, and it can also be applied to other objects.
The program requires 8 bit grayscale images that follow a specific naming scheme, explained in the first video tutorial.
Since 2018 the MPI-Bremen takes care of new software developments and bug-fixing.
The program is available for free for any non-commercial purpose. The usage of the software is at the user's own risk. There is no warranty whatsoever, and there is no guarantee that the obtained results are correct. For any question, contact: A. Ellrott.
Software download:
Automatic Installation: Execute the ACMEtool3 setup file and follow the instructions. You might need to provide admin rights.
Manually Installation: Unzip and copy the whole ACMEtool3 folder to your local hard disk and double-click the application file 'ACMEtool3_2025-05-30'. Generally no further installation is necessary, as the needed Microsoft Net Framework 4.7.2 is usually already present on most of the current computer.
Reference - For citing ACMEtool please use:
Bennke, Christin M., Greta Reintjes, Martha Schattenhofer, Andreas Ellrott, Jörg Wulf, Michael Zeder, and Bernhard M. Fuchs. 2016. ‘Modification of a High-Throughput Automatic Microbial Cell Enumeration System for Shipboard Analyses’. Applied and Environmental Microbiology 82 (11): 3289–96. https://doi.org/10.1128/AEM.03931-15.
References
- Alonso C., Zeder M., Piccini C., Conde D., Pernthaler J. 2009. Ecophysiological differences of betaproteobacterial populations in two hydrochemically distinct compartments of a subtropical lagoon. Environ. Microbiol. 11:867-876. [PMID:19040452]
- Bennke C.M., Reintjes G., Schattenhofer M., Ellrott A., Wulf J., Zeder M., Fuchs B.M. 2016. Modification of a High-Throughput Automatic Microbial Cell Enumeration System for Shipboard Analyses. Appl. Environ. Microbiol. 82:3289-96. [PMID:27016562]
- Pernthaler J., Pernthaler A., Amann R. 2003. Automated enumeration of groups of marine picoplankton after fluorescence in situ hybridizationMicrobiol. 69:2631-2637. [PMID:12732531]
- Salcher M.M., Pernthaler J., Zeder M., Psenner R., Posch T. 2008. Spatio-temporal niche separation of planktonic Betaproteobacteria in an oligo-mesotrophic lake. Environ. Microbiol. 10:2074-2086. [PMID:18430016]
- Schattenhofer M., Fuchs B.M., Amann R., Zubkov M.V., Tarran G.A., Pernthaler J. 2009. Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ. Microbiol. 11:2078-2093. [PMID:19453607]
- Zeder M., Ellrott A., Amann R. 2011. Automated sample area definition for high-throughput microscopy. Cytometry A. 79:306-310. [PMID:21412981]
- Zeder M., Kohler E., Pernthaler J. 2010. Automated quality assessment of autonomously acquired microscopic images of fluorescently stained bacteria. Cytometry A. 77:76-85. [PMID:19821518]
- Zeder M., Pernthaler J. 2009. Multispot live-image autofocusing for high-throughput microscopy of fluorescently stained bacteria. Cytometry A. 75:781-788. [PMID:19658173]
- Zeder M., Peter S., Shabarova T., Pernthaler J. 2009. A small population of planktonic Flavobacteria with disproportionally high growth during the spring phytoplankton bloom in a prealpine lake. Environ. Microbiol. 11:2676-2686. [PMID:19601962]
- Zeder M., van den Wyngaert S., Köster O., Felder K.M., Pernthaler J. 2010. Automated quantification and sizing of unbranched filamentous cyanobacteria by model-based object-oriented image analysis. Appl. Environ. Microbiol. 76:1615-1622. [PMID:20048059]