- Abteilungen
- Abteilung Molekulare Ökologie
- Molekulare Ökologie
- Fluoreszenz-in-situ-Hybridisierung
Fluoreszenz-in-situ-Hybridisierung
Der rRNA-Ansatz
Der rRNA-Ansatz zur Identifizierung, Quantifizierung und Lokalisierung von bisher nicht kultivierten Bakterien
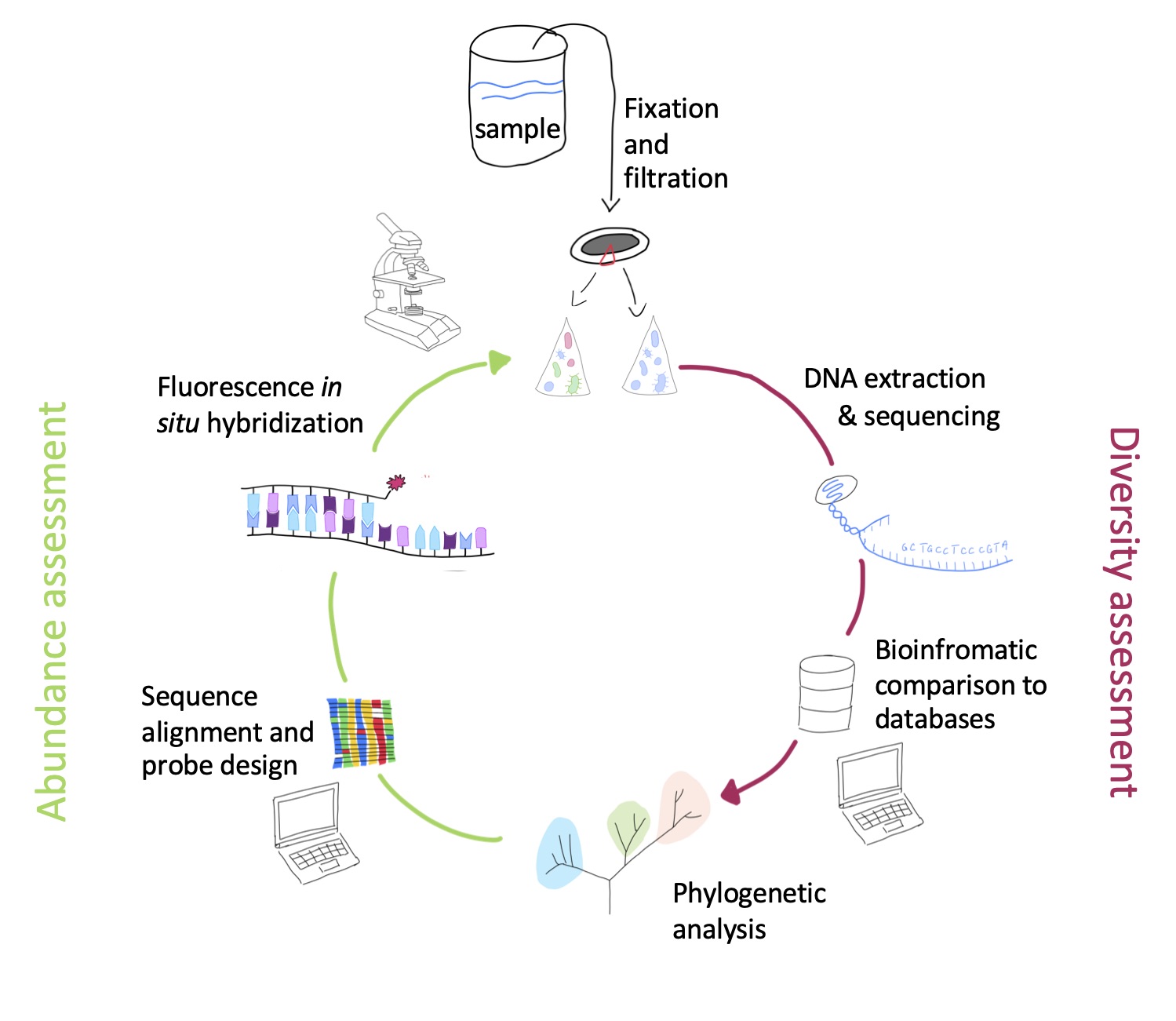
Zur Erforschung von mikrobiellen Lebensgemeinschaften hat sich seit über 30 Jahren der sogenannte ribosomale RNA (rRNA) -Ansatz etabliert. Worauf beruht der rRNA-Ansatz? Im Gegensatz zu mehrzelligen Organismen sind einzellige Mikroben anhand ihrer Morphologie kaum zu unterscheiden. Erschwerend hinzu kommt, dass nur ein kleiner Teil von nicht einmal 1 % aller Umweltmikroben in Kultur gebracht werden können. Deswegen bietet sich die Analyse ihres genetischen Materials an und hier insbesondere der rRNA. Die rRNA ist ein wesentlicher Bestandteil der Ribosomen, den Orten der Proteinbiosynthese. Sie kommt daher in jedem lebenden Organismus vor und hat in jedem Organismus etwa die gleiche Länge. Man nennt sie daher „konserviert“. Ein Vergleich der rRNA-Gensequenzen ermöglicht somit Aufschluss über die Verwandtschaftsbeziehung der Organismen, aus denen die rRNA-Sequenzen stammen. Je weniger Unterschiede in den Sequenzen zu finden sind, desto näher sind die Organismen miteinander verwandt. So ist es möglich, Stammbäume zu rekonstruieren und die Mikroben in eine taxonomische Ordnung zu bringen.
Mit diesem Ansatz können ganze mikrobielle Lebensgemeinschaften erforscht werden, ohne die einzelnen Mitglieder aufwändig in Kultur zu bringen und charakterisieren zu müssen. Außerdem können so neue, noch unbekannte Organismen erkannt werden.
Schematisch sind die Arbeitsschritte in der Abbildung rechts beschrieben: Im Labor isoliert man die gesamte DNA aus der Umweltprobe und amplifiziert die Genabschnitte, die für die rRNA kodieren, mit spezifischen Primern mittels Polymerase-Ketten-Reaktion (PCR). Im nächsten Schritt werden die Produkte aus der PCR sequenziert und die resultierenden Sequenzen der rRNA Gene mit den bereits bekannten Sequenzen aus öffentlichen Datenbanken verglichen. So kann die Diversität der in der Probe lebenden Mikroben umfassend bestimmt werden. Im nächsten Schritt geht es um die Visualisierung und Quantifizierung der Mikroben mit der Fluoreszenz-in-situ-Hybridisierung, die im nächsten Abschnitt beschrieben sind.
Fluoreszenz-in-situ-Hybridisierung
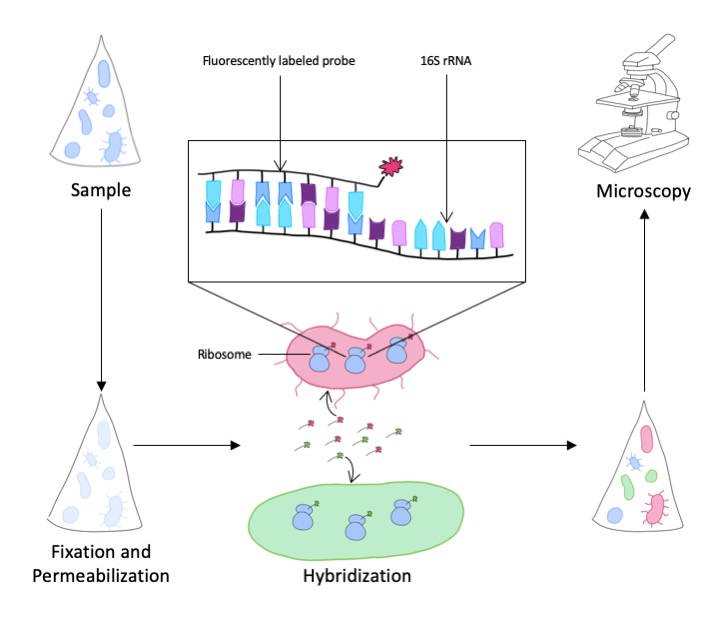
Die Sequenzinformation aus der rRNA sagt jedoch noch nichts Genaues über die Häufigkeit einer bestimmten Art oder Gattung in der Umweltprobe aus. Dazu verwenden wir eine weitere Methode, die es ermöglicht, die Mikroben direkt in der Probe anzufärben, damit zu visualisieren und dann zu zählen. Im ersten Schritt werden Signaturen in der rRNA gesucht, die spezifisch für die taxonomische Gruppe der Mikroorganismen sind, die man untersuchen will. Gegen diese Signaturen werden komplementäre, kurze DNA-Sequenzen synthetisiert, die mit einem Fluoreszenzfarbstoff markiert sind. Diese sogenannten Sonden binden in einer Umweltprobe unter geeigneten experimentellen Bedingungen nur an die rRNA der Zielorganismen, aber nicht an den Rest der vorhandenen Zellen. Dadurch ist es möglich, nicht nur die Zellen in der Probe anzufärben und zu quantifizieren, sondern auch zu lokalisieren. Oft leben Mikroben in Konsortien oder angeheftet auf Oberflächen, die so nun in ihrem natürlichen Umfeld visualisiert werden können. Eine wichtige Voraussetzung dafür ist, dass Umweltproben möglichst zeitnah nach ihrer Beprobung mit einem Fixativ (z. B. Formaldehyd) haltbar gemacht werden, um die natürliche Lebensgemeinschaft bestmöglich in Form und Verteilung zu erhalten.
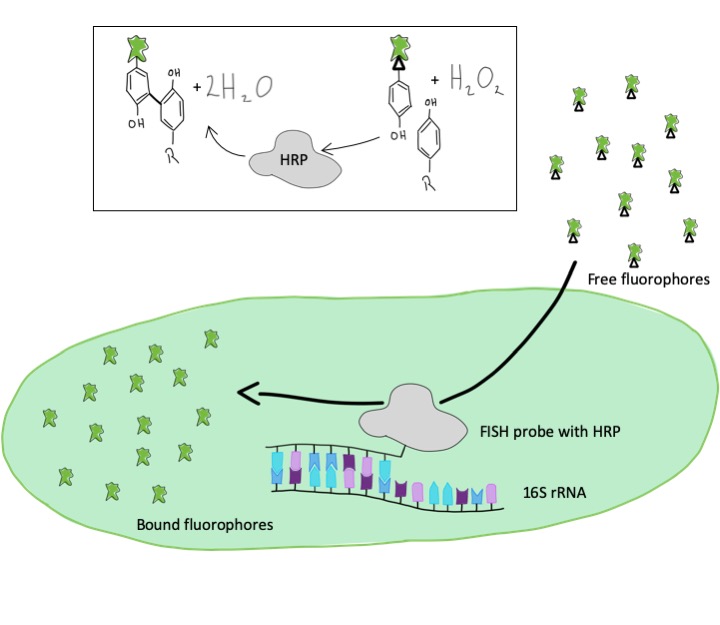
Catalyzed Reporter Decomposition - ein stärkeres Signal mit CARD-FISH
Um ein stärkeres Signal und heller leuchtende Zellen zu bekommen, behelfen wir uns mit einer enzymatischen Reaktion. Anstelle der direkten Markierung mit einem Fluoreszenzfarbstoff nutzen wir DNA-Fragmente mit einem Enzym, einer sogenannten Peroxidase. In einem zweiten Schritt werden nun die Farbstoffmoleküle zu den Zellen gegeben und die Peroxidase bindet diese Farbstoffmoleküle an umliegende Proteine. Somit bekommen wir ein Signal, was bis zu 50-fach stärker ist als bei der direkten Markierung. Wir können so auch Mikroorganismen sichtbar machen, die besonders wenig rRNA-Sequenzen enthalten und die wir mit der herkömmlichen Methode nicht detektieren konnten.
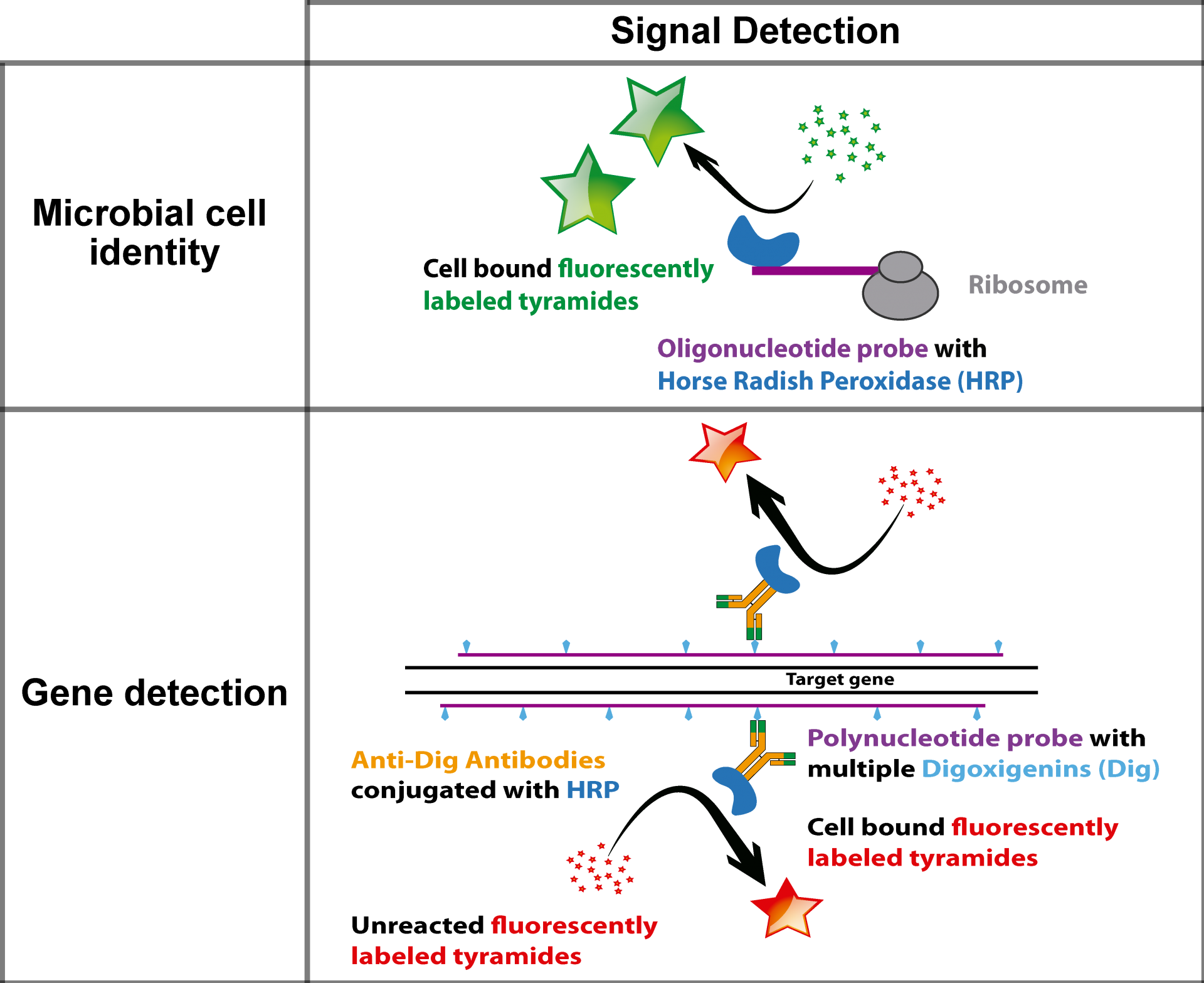
geneFISH
Nachweis und Visualisierung einzelner Gene in Bakterienzellen
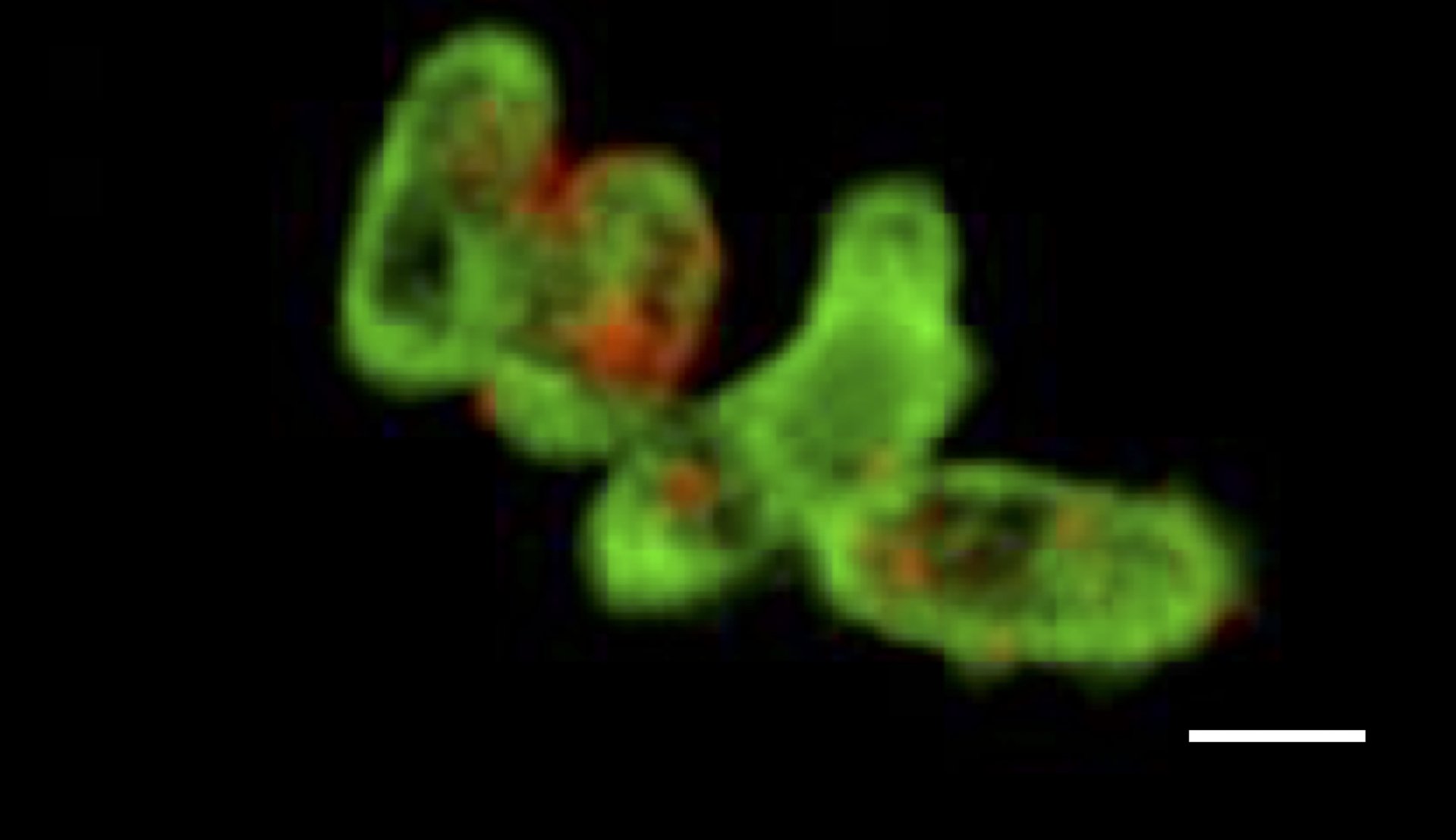
GeneFISH kombiniert den Nachweis von spezifischen Genen und ribosomaler RNA (rRNA) auf der Ebene einzelner Zellen. Während bestimmte Gene auf eine bestimmte funktionelle Eigenschaft hinweisen, erlaubt die rRNA die taxonomische Zugehörigkeit der Mikroorganismen. Mit geneFISH können wir also eine mögliche Stoffwechselfunktion direkt mit der mikrobiellen Taxonomie verknüpfen. Wir synthetisieren Gen-spezifische dsDNA-Polynukleotid-Sonden, die mit vielen Digoxigeninen markiert sind, die wiederum als Ziel von Peroxidase-markierten Antikörpern dienen. In einer CARD Reaktion (siehe CARD-FISH) wird das Gen schließlich markiert. Alternativ sind die Polynukleotid-Sonden direkt mit mehreren Fluoreszenzfarbstoffen markiert (direct-GeneFISH). Üblicherweise kombinieren wir 16S rRNA FISH mit geneFISH, um die Mikroorganismen zu identifizieren und gleichzeitig Schlüsselgene nachzuweisen.
Lesen Sie mehr über geneFISH in dieser Originalpublikation: GeneFISH – an in situ technique for linking gene presence and cell identity in environmental microorganisms
oder über Direct-geneFISH: Direct-geneFISH: a simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms
Identifizierung phageninfizierter Zellen mittels phageFISH
Phagen sind Viren, die bakterielle und archaeale Zellen befallen. Die geneFISH-Methode wurde zum Nachweis einzelner Phagengene erweitert. Damit können wir sowohl Mikroorganismen identifizieren, die von einem Virus infiziert wurden, als auch frei schwimmende Phagen.
Lesen Sie mehr über phageFISH in dieser Originalpublikation: Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses.